| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 21,252,975 – 21,253,298 |
| Length | 323 |
| Max. P | 0.998011 |

| Location | 21,252,975 – 21,253,085 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 92.96 |
| Mean single sequence MFE | -24.82 |
| Consensus MFE | -18.91 |
| Energy contribution | -19.75 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.640793 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 21252975 110 + 27905053 UGAACUCGAUCUUUCCUCAAAUUGCUUUCCGCCUCG-UUUUUAUAACCCUUUCAACUCUGUAGAUUUAUGUUCCAUU-UUAGGUGGCGCCACCUGGCGUCGCCUACCUCUGG .((((..(((((...........((.....))...(-((.....)))..............)))))...))))....-.(((((((((((....)))))))))))....... ( -25.60) >DroSec_CAF1 78061 110 + 1 UGAACUCGAUCUUUCCUCAAAUUGCUUUCCGCCUCG-UUUUUAUAACCCUUUCAACUCUGUAGAUUUAUGUUACAUU-UUAGGUGGCGCCACCUGGCGUCGCCUACCUCUGG (((((..(((((...........((.....))...(-((.....)))..............)))))...))).))..-.(((((((((((....)))))))))))....... ( -23.20) >DroSim_CAF1 80750 110 + 1 UGAACUCGAUCUUUCCUCAAAUUGCUUUCCGCCUCG-UUUUUAUAACCCUUUCAACUCUGUAGAUUUAUGUUCCAUU-UUAGGUGGCGCCACCUGGCGUCGCCUACCUCCGG .((((..(((((...........((.....))...(-((.....)))..............)))))...))))....-.(((((((((((....)))))))))))....... ( -25.60) >DroYak_CAF1 87624 110 + 1 UGAACUCGAUCUUUCCUCAAAUUACUUUCCGCCUCG-UUUUUAUAACCCUUUCAACUCCGUAGAUUUAUGUUCCACU-UUAGGUGGCGCCACCUGGCGUCGCCUACCUCUGG .((((..(((((........................-........................)))))...))))....-.(((((((((((....)))))))))))....... ( -23.61) >DroAna_CAF1 72550 112 + 1 UGAACUGGCUCUCGCCCCAAAUUGGUUUCCGCCUCGCUUUUUAUAACCCUUUCAACUUUGUGGAUUUAUGUUCCAUUUUUAGGUGGCGCCACCUGGCCUCGCAUUCCGCCGG ....(((((....(((((.....(((....)))..........................(((((.......))))).....)).)))(((....)))..........))))) ( -26.10) >consensus UGAACUCGAUCUUUCCUCAAAUUGCUUUCCGCCUCG_UUUUUAUAACCCUUUCAACUCUGUAGAUUUAUGUUCCAUU_UUAGGUGGCGCCACCUGGCGUCGCCUACCUCUGG .((((..(((((.................................................)))))...))))......(((((((((((....)))))))))))....... (-18.91 = -19.75 + 0.84)



| Location | 21,253,014 – 21,253,111 |
|---|---|
| Length | 97 |
| Sequences | 5 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 92.61 |
| Mean single sequence MFE | -23.38 |
| Consensus MFE | -19.66 |
| Energy contribution | -20.90 |
| Covariance contribution | 1.24 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.62 |
| Structure conservation index | 0.84 |
| SVM decision value | 2.98 |
| SVM RNA-class probability | 0.998011 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 21253014 97 + 27905053 UUAUAACCCUUUCAACUCUGUAGAUUUAUGUUCCAUU-UUAGGUGGCGCCACCUGGCGUCGCCUACCUCUGGCAAUAAAAUAUAUAUACCAAUGCUAC ..................((((.((((.(((((((..-.(((((((((((....)))))))))))....))).)))))))).))))............ ( -25.30) >DroSec_CAF1 78100 97 + 1 UUAUAACCCUUUCAACUCUGUAGAUUUAUGUUACAUU-UUAGGUGGCGCCACCUGGCGUCGCCUACCUCUGGCAAUAAAAUAUAUAUACCAAUGCUAC ..................((((..(((((((((....-.(((((((((((....)))))))))))....)))).))))).)))).............. ( -23.50) >DroSim_CAF1 80789 97 + 1 UUAUAACCCUUUCAACUCUGUAGAUUUAUGUUCCAUU-UUAGGUGGCGCCACCUGGCGUCGCCUACCUCCGGCAAUAAAAUAUAUAUACCAAUGCUAC ..................((((.((((.((((((...-.(((((((((((....))))))))))).....)).)))))))).))))............ ( -23.70) >DroYak_CAF1 87663 97 + 1 UUAUAACCCUUUCAACUCCGUAGAUUUAUGUUCCACU-UUAGGUGGCGCCACCUGGCGUCGCCUACCUCUGGCAAUAAAAUAUAUAUAGCAAUGCUCC ...................(((.((((.(((((((..-.(((((((((((....)))))))))))....))).)))))))).)))............. ( -24.60) >DroAna_CAF1 72590 98 + 1 UUAUAACCCUUUCAACUUUGUGGAUUUAUGUUCCAUUUUUAGGUGGCGCCACCUGGCCUCGCAUUCCGCCGGCAAUAAAAUAUAUAUGCGAUUGCACC ...................(((((.......)))))...((((((....))))))(((..((.....)).))).............(((....))).. ( -19.80) >consensus UUAUAACCCUUUCAACUCUGUAGAUUUAUGUUCCAUU_UUAGGUGGCGCCACCUGGCGUCGCCUACCUCUGGCAAUAAAAUAUAUAUACCAAUGCUAC ..................((((.((((.(((((((....(((((((((((....)))))))))))....))).)))))))).))))............ (-19.66 = -20.90 + 1.24)


| Location | 21,253,050 – 21,253,145 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 86.91 |
| Mean single sequence MFE | -23.83 |
| Consensus MFE | -17.31 |
| Energy contribution | -18.11 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.05 |
| Structure conservation index | 0.73 |
| SVM decision value | 1.87 |
| SVM RNA-class probability | 0.980839 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 21253050 95 + 27905053 U-UUAGGUGGCGCCACCUGGCGUCGCCUACCUCUGGCAAUAAAAUAUAUAUACCAAUGCUACU--AUA----UAUAAAGGCGAACAAUAAAAACACAACUUC .-.(((((((((((....)))))))))))......((.......((((((((..........)--)))----))))...))..................... ( -23.30) >DroSec_CAF1 78136 95 + 1 U-UUAGGUGGCGCCACCUGGCGUCGCCUACCUCUGGCAAUAAAAUAUAUAUACCAAUGCUACU--AUA----UAUAAAGGCGAACAAUAAAAACACAACUUC .-.(((((((((((....)))))))))))......((.......((((((((..........)--)))----))))...))..................... ( -23.30) >DroSim_CAF1 80825 95 + 1 U-UUAGGUGGCGCCACCUGGCGUCGCCUACCUCCGGCAAUAAAAUAUAUAUACCAAUGCUACU--AUA----UAUAAAGGCGAACAAUAAAAACACAACUUC .-.(((((((((((....)))))))))))......((.......((((((((..........)--)))----))))...))..................... ( -23.30) >DroYak_CAF1 87699 99 + 1 U-UUAGGUGGCGCCACCUGGCGUCGCCUACCUCUGGCAAUAAAAUAUAUAUAGCAAUGCUCCU--AUAUAUAUAUAAAGGCGAACAAUAAAAACACAACUUC .-.(((((((((((....)))))))))))......((......((((((((((........))--))))))))......))..................... ( -29.70) >DroAna_CAF1 72626 98 + 1 UUUUAGGUGGCGCCACCUGGCCUCGCAUUCCGCCGGCAAUAAAAUAUAUAUGCGAUUGCACCCCGCCA----UAUCCCGGAGAACAACAAAAGCAAUAUUCC ...((((((....))))))(((..((.....)).))).............(((..(((.......((.----......)).......)))..)))....... ( -19.54) >consensus U_UUAGGUGGCGCCACCUGGCGUCGCCUACCUCUGGCAAUAAAAUAUAUAUACCAAUGCUACU__AUA____UAUAAAGGCGAACAAUAAAAACACAACUUC ...(((((((((((....))))))))))).....((((..................)))).......................................... (-17.31 = -18.11 + 0.80)


| Location | 21,253,111 – 21,253,219 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 88.68 |
| Mean single sequence MFE | -17.78 |
| Consensus MFE | -15.00 |
| Energy contribution | -15.08 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.41 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.625604 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 21253111 108 + 27905053 U--AUA----UAUAAAGGCGAACAAUAAAAACACAACUUCAUACACGACAUAAAUCAACCGGGCAGUUCAAGCAUGGCUGAAAUAUAACUCAGCCGAGAAGUUAAAUACAAACC .--...----........................((((((......................((.......)).(((((((........))))))).))))))........... ( -17.20) >DroSec_CAF1 78197 108 + 1 U--AUA----UAUAAAGGCGAACAAUAAAAACACAACUUCAUACACGACAUAAAUCAACCGGGCAGUUCAAGCAUGGCUGAAAUAUAACUCAGCCGAGAAGUUAAAUACAAACC .--...----........................((((((......................((.......)).(((((((........))))))).))))))........... ( -17.20) >DroSim_CAF1 80886 108 + 1 U--AUA----UAUAAAGGCGAACAAUAAAAACACAACUUCAUACACGACAUAAAUCAACCGGGCAGUUCAAGCAUGGCUGAAAUAUAACUCAGCCGAGAAGUUAAAUACAAACC .--...----........................((((((......................((.......)).(((((((........))))))).))))))........... ( -17.20) >DroYak_CAF1 87760 112 + 1 U--AUAUAUAUAUAAAGGCGAACAAUAAAAACACAACUUCAUACACGACAUAAAUCAACCGGGCAGUUCAAGCAUGGCUGAAAUAUUACUCAGCCGAGAAGUUAAAUACAAUCU .--...............................((((((......................((.......)).(((((((........))))))).))))))........... ( -17.20) >DroAna_CAF1 72688 110 + 1 CCGCCA----UAUCCCGGAGAACAACAAAAGCAAUAUUCCAUGCACGACAUAAAUCACCUGGGCAGUUCAAGCCGGGCUGAAUUAUUACUCGGCCGAGAAGUUAAAUAAAAGCC ......----..((.(((............(((........))).(((.((((.(((.((.(((.......))).)).))).))))...))).))).))............... ( -20.10) >consensus U__AUA____UAUAAAGGCGAACAAUAAAAACACAACUUCAUACACGACAUAAAUCAACCGGGCAGUUCAAGCAUGGCUGAAAUAUAACUCAGCCGAGAAGUUAAAUACAAACC ..................................((((((......................((.......))..((((((........))))))..))))))........... (-15.00 = -15.08 + 0.08)


| Location | 21,253,179 – 21,253,298 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.65 |
| Mean single sequence MFE | -30.35 |
| Consensus MFE | -26.65 |
| Energy contribution | -26.90 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.706700 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 21253179 119 + 27905053 UGGCUGAAAUAUAACUCAGCCGAGAAGUUAAAUACAAACCGCCACCAGCUGACAGCAA-AAAAUGGCAAAGGGCGUGGUCAAUUAGAGUAAAAGUUGGGUGUAAGUAAUGCCCAGAUUGU .((((((........))))))............((((...(((((..(((....((..-......))....))))))))...............((((((((.....)))))))).)))) ( -28.00) >DroSec_CAF1 78265 119 + 1 UGGCUGAAAUAUAACUCAGCCGAGAAGUUAAAUACAAACCGCCACCCACUGACAGAAA-AAAAUGGCAAAGGGCGUGGUCAAUUAGAGUAAAAGUUGGGUGUAAGUAAUGCCCAGAUUGU .((((((........))))))............(((((((((..(((..((.((....-....)).))..))).)))))...............((((((((.....)))))))).)))) ( -30.20) >DroSim_CAF1 80954 119 + 1 UGGCUGAAAUAUAACUCAGCCGAGAAGUUAAAUACAAACCGCCACCCACUGACAGCAA-AAAAUGGCAAAGGGCGUGGUCAAUUAGAGUAAAAGUUGGGUGUAAGUAAUGCCCAGAUUGU .((((((........))))))............(((((((((..(((..((.((....-....)).))..))).)))))...............((((((((.....)))))))).)))) ( -30.20) >DroYak_CAF1 87832 120 + 1 UGGCUGAAAUAUUACUCAGCCGAGAAGUUAAAUACAAUCUGCCACCCACUGACAACAAAAAAAUGGCAAAGGGCGUGGUCAAUUAGAGUAAAAGUUGGGUGUAAGUAAUGCCCAGAUUGU .((((((........))))))............((((((((((((((.((.....((......))....)))).))))).................((((((.....))))))))))))) ( -33.00) >consensus UGGCUGAAAUAUAACUCAGCCGAGAAGUUAAAUACAAACCGCCACCCACUGACAGCAA_AAAAUGGCAAAGGGCGUGGUCAAUUAGAGUAAAAGUUGGGUGUAAGUAAUGCCCAGAUUGU .((((((........))))))............((((...(((((((..((.((.........)).))...)).)))))...............((((((((.....)))))))).)))) (-26.65 = -26.90 + 0.25)

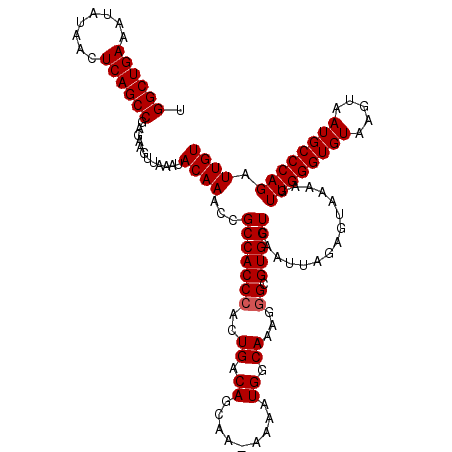

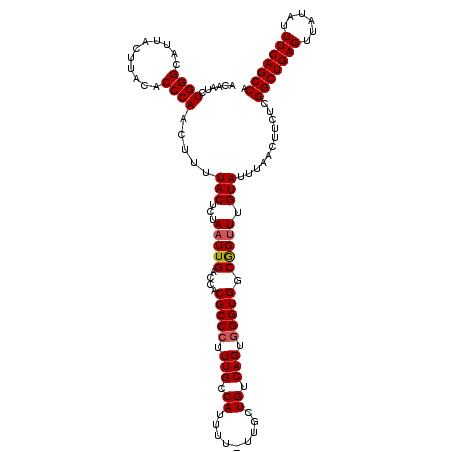
| Location | 21,253,179 – 21,253,298 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.65 |
| Mean single sequence MFE | -30.12 |
| Consensus MFE | -26.34 |
| Energy contribution | -26.65 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.47 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.95 |
| SVM RNA-class probability | 0.888232 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 21253179 119 - 27905053 ACAAUCUGGGCAUUACUUACACCCAACUUUUACUCUAAUUGACCACGCCCUUUGCCAUUUU-UUGCUGUCAGCUGGUGGCGGUUUGUAUUUAACUUCUCGGCUGAGUUAUAUUUCAGCCA ((((.((((((...................................))))...((((((..-..((.....)).)))))))).))))............(((((((......))))))). ( -28.15) >DroSec_CAF1 78265 119 - 1 ACAAUCUGGGCAUUACUUACACCCAACUUUUACUCUAAUUGACCACGCCCUUUGCCAUUUU-UUUCUGUCAGUGGGUGGCGGUUUGUAUUUAACUUCUCGGCUGAGUUAUAUUUCAGCCA ......((((...........))))............((.((((.(((((.(((.((....-....)).))).)))))..)))).))............(((((((......))))))). ( -30.10) >DroSim_CAF1 80954 119 - 1 ACAAUCUGGGCAUUACUUACACCCAACUUUUACUCUAAUUGACCACGCCCUUUGCCAUUUU-UUGCUGUCAGUGGGUGGCGGUUUGUAUUUAACUUCUCGGCUGAGUUAUAUUUCAGCCA ......((((...........))))............((.((((.(((((.(((.((....-....)).))).)))))..)))).))............(((((((......))))))). ( -30.10) >DroYak_CAF1 87832 120 - 1 ACAAUCUGGGCAUUACUUACACCCAACUUUUACUCUAAUUGACCACGCCCUUUGCCAUUUUUUUGUUGUCAGUGGGUGGCAGAUUGUAUUUAACUUCUCGGCUGAGUAAUAUUUCAGCCA (((((((((((...................................))))...(((((((..(((....))).))))))))))))))............(((((((......))))))). ( -32.15) >consensus ACAAUCUGGGCAUUACUUACACCCAACUUUUACUCUAAUUGACCACGCCCUUUGCCAUUUU_UUGCUGUCAGUGGGUGGCGGUUUGUAUUUAACUUCUCGGCUGAGUUAUAUUUCAGCCA ......((((...........)))).....(((...(((((....(((((.(((.((.........)).))).))))).))))).)))...........(((((((......))))))). (-26.34 = -26.65 + 0.31)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:58:15 2006