
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 21,244,273 – 21,244,365 |
| Length | 92 |
| Max. P | 0.711708 |

| Location | 21,244,273 – 21,244,365 |
|---|---|
| Length | 92 |
| Sequences | 4 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 83.25 |
| Mean single sequence MFE | -19.56 |
| Consensus MFE | -12.82 |
| Energy contribution | -14.88 |
| Covariance contribution | 2.06 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.38 |
| SVM RNA-class probability | 0.711708 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
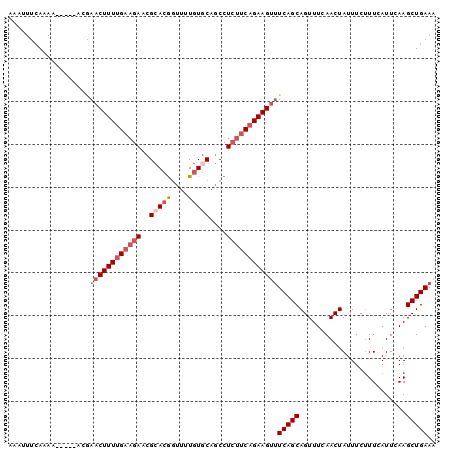
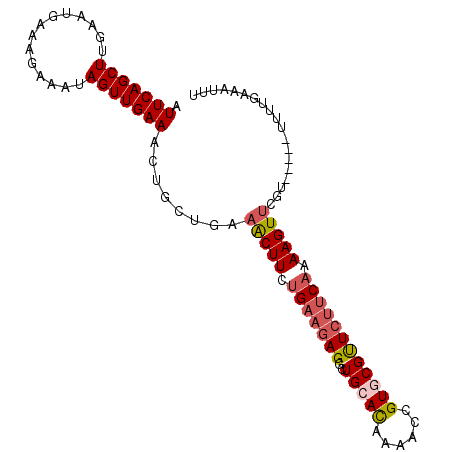
>3R_DroMel_CAF1 21244273 92 + 27905053 AAAUUUCAAAA-----ACAAACUUUUGAAGAACGCACGGUUUUGUGCAGCCU---CAGAAGUAUCAGCAGUUUCAACUAUUUCGUUCAUUGAAGCUGAAU ...........-----....((((((((.(...(((((....)))))..).)---))))))).....(((((((((..(......)..)))))))))... ( -21.70) >DroSec_CAF1 69033 95 + 1 AAAUUUCAAAA-----AUGAACUUUUGAAGAACGCACGGUUUUGUUCAGCCUCUUCAGAAGUUUCAGCAGUUUCAACUAUUUCUUUUAUUCAAGCUGAAA ...........-----...((((((((((((......((((......))))))))))))))))(((((.........................))))).. ( -18.61) >DroSim_CAF1 71340 95 + 1 AAAUUUCAAAA-----ACGAACUUUUGAAGAACGCACGGUUUUGUGCAGCCUCUUCAGAAGUUUCAGCAGUUUCAACUAUUUCUUUCAUUCAAGCUGAAA ...........-----...((((((((((((..(((((....)))))....))))))))))))(((((.........................))))).. ( -22.21) >DroEre_CAF1 73087 96 + 1 ----UGCAAAAUAUUUGCCAACUUUUGAAGAGCGAACAAAUUUAUGCAACCUCUUCUGAAGCUUCAGCAGUUUCAACUAUUUCUUUAAUUCAAGCUGAAU ----.((((.....))))...((((.((((((....((......))....)))))).)))).((((((.........................)))))). ( -15.71) >consensus AAAUUUCAAAA_____ACGAACUUUUGAAGAACGCACGGUUUUGUGCAGCCUCUUCAGAAGUUUCAGCAGUUUCAACUAUUUCUUUCAUUCAAGCUGAAA ...................((((((((((((..(((((....)))))....))))))))))))(((((.........................))))).. (-12.82 = -14.88 + 2.06)


| Location | 21,244,273 – 21,244,365 |
|---|---|
| Length | 92 |
| Sequences | 4 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 83.25 |
| Mean single sequence MFE | -21.39 |
| Consensus MFE | -14.75 |
| Energy contribution | -15.94 |
| Covariance contribution | 1.19 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.31 |
| SVM RNA-class probability | 0.681372 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
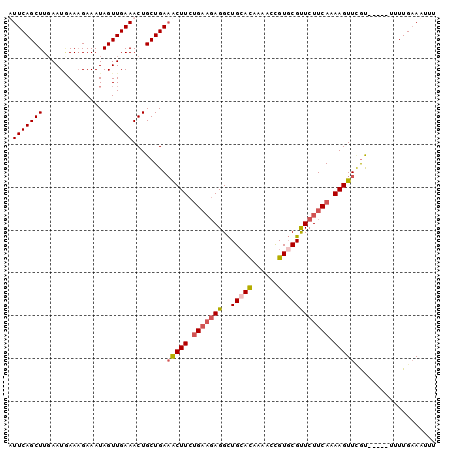
>3R_DroMel_CAF1 21244273 92 - 27905053 AUUCAGCUUCAAUGAACGAAAUAGUUGAAACUGCUGAUACUUCUG---AGGCUGCACAAAACCGUGCGUUCUUCAAAAGUUUGU-----UUUUGAAAUUU ..(((((((((((..........))))))...))))).((((.((---(((.(((((......)))))..))))).))))....-----........... ( -21.30) >DroSec_CAF1 69033 95 - 1 UUUCAGCUUGAAUAAAAGAAAUAGUUGAAACUGCUGAAACUUCUGAAGAGGCUGAACAAAACCGUGCGUUCUUCAAAAGUUCAU-----UUUUGAAAUUU ((((((((..............))))))))....((((.(((.(((((((((((........)).)).))))))).))))))).-----........... ( -18.54) >DroSim_CAF1 71340 95 - 1 UUUCAGCUUGAAUGAAAGAAAUAGUUGAAACUGCUGAAACUUCUGAAGAGGCUGCACAAAACCGUGCGUUCUUCAAAAGUUCGU-----UUUUGAAAUUU ((((((...((((((......((((.......))))..((((.(((((((..(((((......)))))))))))).))))))))-----))))))))... ( -22.50) >DroEre_CAF1 73087 96 - 1 AUUCAGCUUGAAUUAAAGAAAUAGUUGAAACUGCUGAAGCUUCAGAAGAGGUUGCAUAAAUUUGUUCGCUCUUCAAAAGUUGGCAAAUAUUUUGCA---- ..(((((((.(((((......))))).))...)))))(((((..((((((...(((......)))...))))))..))))).((((.....)))).---- ( -23.20) >consensus AUUCAGCUUGAAUGAAAGAAAUAGUUGAAACUGCUGAAACUUCUGAAGAGGCUGCACAAAACCGUGCGUUCUUCAAAAGUUCGU_____UUUUGAAAUUU .(((((((..............)))))))........(((((.(((((((..(((((......)))))))))))).)))))................... (-14.75 = -15.94 + 1.19)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:58:02 2006