
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 21,238,485 – 21,238,641 |
| Length | 156 |
| Max. P | 0.981939 |

| Location | 21,238,485 – 21,238,601 |
|---|---|
| Length | 116 |
| Sequences | 3 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 92.08 |
| Mean single sequence MFE | -35.27 |
| Consensus MFE | -35.24 |
| Energy contribution | -34.80 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.22 |
| Structure conservation index | 1.00 |
| SVM decision value | 1.90 |
| SVM RNA-class probability | 0.981939 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
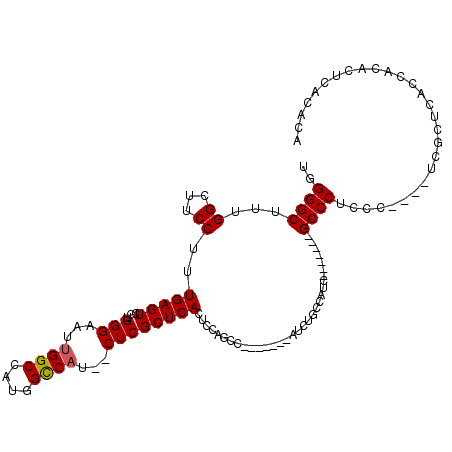
>3R_DroMel_CAF1 21238485 116 + 27905053 UUUACGUUUUUGUCUUCAAGAGCCUCGAGGGCGUUUUUCUGGGGCUUUGGCUUCCUUUGAGUCCUGGGAAUUGGCCAUGGCCAUCUCGCUCACUCCAGCCAUCUCUCAUCUCCCAU .....(((((((....))))))).....(((((..((((..((((((.((...))...))))))..)))).((((....))))...)))))......................... ( -33.30) >DroSim_CAF1 65441 109 + 1 UUUACGCUUUUGUCUUCAAGAGCCUCGAGGGCGUUUUUCUGGGGCUUUGGCUUCCUUUGAGUCCUGGGAAUUGGCCAUGGCCAUCUCGCUCACUCCACCC-------AUCUACCAU .....(((((((....))))))).....(((((..((((..((((((.((...))...))))))..)))).((((....))))...))))).........-------......... ( -35.70) >DroEre_CAF1 67268 109 + 1 UUUACGCUUUUGUCUUCGAGAGCCUCGAGGGCGUUUUUCUGGGGCUUUGGCUUCCUUUGAGUCCUGGGAAUUGGCCAUGGCCAUCUCGCUCACUCCAGCU-------AUUUGCCAU ...........(((((((((...)))))))))..........(((..(((((.....(((((...((((..((((....)))))))))))))....))))-------)...))).. ( -36.80) >consensus UUUACGCUUUUGUCUUCAAGAGCCUCGAGGGCGUUUUUCUGGGGCUUUGGCUUCCUUUGAGUCCUGGGAAUUGGCCAUGGCCAUCUCGCUCACUCCAGCC_______AUCUACCAU .....(((((((....))))))).....(((((..((((..((((((.((...))...))))))..)))).((((....))))...)))))......................... (-35.24 = -34.80 + -0.44)



| Location | 21,238,524 – 21,238,641 |
|---|---|
| Length | 117 |
| Sequences | 4 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 82.54 |
| Mean single sequence MFE | -26.40 |
| Consensus MFE | -18.29 |
| Energy contribution | -18.35 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.69 |
| SVM decision value | -0.04 |
| SVM RNA-class probability | 0.514378 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 21238524 117 + 27905053 UGGGGCUUUGGCUUCCUUUGAGUCCUGGGAAUUGGCCAUGGCCAU--CUCGCUCACUCCAGCCAUCUCUCAUCUCCCAUCGCCUAUUGCCCACCCCCCGUCGCUCACCACACUCACACA .((((...(((((.....(((((...((((..((((....)))))--))))))))....)))))................((.....)).....))))..................... ( -28.00) >DroSim_CAF1 65480 110 + 1 UGGGGCUUUGGCUUCCUUUGAGUCCUGGGAAUUGGCCAUGGCCAU--CUCGCUCACUCCACCC-------AUCUACCAUCGCCUAUCGCCCUCCCGCCGUCGCUCACCACACACACACA ..((((..((((......(((((...((((..((((....)))))--))))))))........-------..........((.....))......))))..)))).............. ( -26.40) >DroEre_CAF1 67307 99 + 1 UGGGGCUUUGGCUUCCUUUGAGUCCUGGGAAUUGGCCAUGGCCAU--CUCGCUCACUCCAGCU-------AUUUGCCAUC-------GCCCUCCC----ACGCUCACCACACUCACACA .(((((....)))))...(((((..(((((...(((.(((((...--...(((......))).-------....))))).-------))).))))----).)))))............. ( -30.10) >DroYak_CAF1 72530 101 + 1 UGGGGCUUUGGCUUCCUUUGAGUCCUGGGAAUUGGCCAUCGUCCUCUCUCGCUCACUCCAUUC-------AUCUGCUAUC-------GCCCUCCC----UCGCUCAGCACACCCACACA .(((((..((((.......((((..(((((...(((....)))...)))))...)))).....-------....))))..-------)))))...----.................... ( -21.09) >consensus UGGGGCUUUGGCUUCCUUUGAGUCCUGGGAAUUGGCCAUGGCCAU__CUCGCUCACUCCAGCC_______AUCUGCCAUC_______GCCCUCCC____UCGCUCACCACACUCACACA ..((((...((...))..(((((...(((...((((....))))...))))))))................................))))............................ (-18.29 = -18.35 + 0.06)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:58:00 2006