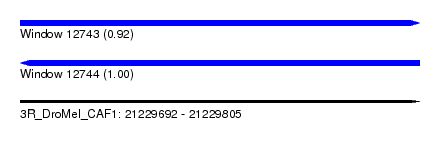
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 21,229,692 – 21,229,805 |
| Length | 113 |
| Max. P | 0.999144 |

| Location | 21,229,692 – 21,229,805 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 72.93 |
| Mean single sequence MFE | -35.01 |
| Consensus MFE | -14.38 |
| Energy contribution | -15.94 |
| Covariance contribution | 1.56 |
| Combinations/Pair | 1.35 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.41 |
| SVM decision value | 1.15 |
| SVM RNA-class probability | 0.923451 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
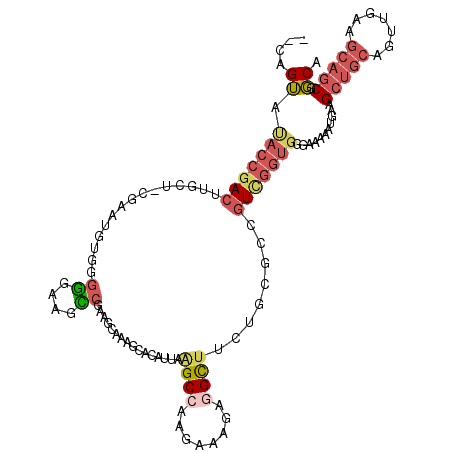
>3R_DroMel_CAF1 21229692 113 + 27905053 --CAGUACACCGACUUGCU-CGAAUGUGGGGGAAGCCGAAGCAAAGCACAUUAAGCCAAGAAAGAGGCUUCUGCGGAGUCGGUGGGAAAAAUGGAGCUGCAGUUGAAGCAGCGGCA --..((.(((((((((((.-.....)).(.(((((((........((.......)).........))))))).).)))))))))...........(((((.......))))).)). ( -37.63) >DroSim_CAF1 56608 113 + 1 --CAGUAUACCGACUUGCU-CGACUGUGGGGGAAGCCGAAGCAAAGCACAUUAAGCGAAGAAAGAGGCUUCUGUGGCGUUGGUGGGAAAAAUGGAGCUGCAGUUGAAGCAGCGGCA --.......((..((..(.-((((.((.(.(((((((........((.......)).........))))))).).)))))))..)).........(((((.......))))))).. ( -35.83) >DroEre_CAF1 57888 114 + 1 -CCAGUAUACCGACUUGCU-GGAAUGUGGGGGAAGUCGAAUCAAAGCACAUUAAGCCAAGAAAGAGGCCUCUGCGCCGUCGGUGGGAAAAAUGAAGCUGCAGUUGAAGCAGCGGCA -((....(((((((..(((-..((((((...((.......))....)))))).))).........(((......))))))))))...........(((((.......))))))).. ( -32.70) >DroYak_CAF1 58872 100 + 1 ACCGGUAUACCGACUUUCU-CUAAUGUGGGAGAAGU---------------UAAGCCAAGAAAGAGGCUCCUGCGCCGUCGGUGGGAAAAAUGAAGCUGCAGUUGAAGCAGCGGCA .((....(((((((.((((-((......))))))((---------------..((((........))))...))...)))))))...........(((((.......))))))).. ( -32.80) >DroAna_CAF1 50684 92 + 1 ----GGGCGGCGA--UGCUGCGACGAU--GGGAAGCCACAUCGAAACACAUUAGGCCAAGGAAGCUGUUGC-----CAU-----GAAAAAAUGAAG------UUGCAGCAGCGCCA ----....((((.--((((((((((((--(((...)).))))...........(((..((....))...))-----)..-----...........)------)))))))).)))). ( -36.10) >consensus __CAGUAUACCGACUUGCU_CGAAUGUGGGGGAAGCCGAAGCAAAGCACAUUAAGCCAAGAAAGAGGCUUCUGCGCCGUCGGUGGGAAAAAUGAAGCUGCAGUUGAAGCAGCGGCA ....((.(((((((...............((....))................((((........))))........)))))))...........(((((.......))))).)). (-14.38 = -15.94 + 1.56)


| Location | 21,229,692 – 21,229,805 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 72.93 |
| Mean single sequence MFE | -32.72 |
| Consensus MFE | -12.54 |
| Energy contribution | -13.38 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.36 |
| Mean z-score | -2.81 |
| Structure conservation index | 0.38 |
| SVM decision value | 3.40 |
| SVM RNA-class probability | 0.999144 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
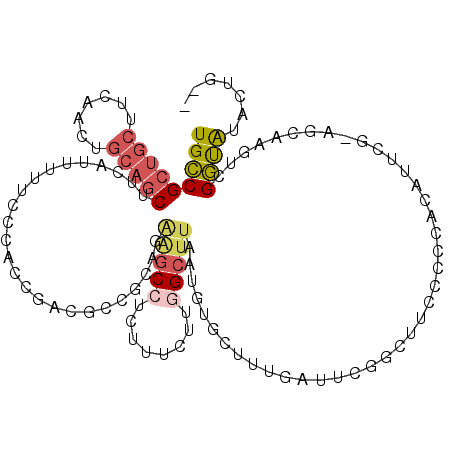
>3R_DroMel_CAF1 21229692 113 - 27905053 UGCCGCUGCUUCAACUGCAGCUCCAUUUUUCCCACCGACUCCGCAGAAGCCUCUUUCUUGGCUUAAUGUGCUUUGCUUCGGCUUCCCCCACAUUCG-AGCAAGUCGGUGUACUG-- ....(((((.......)))))...........((((((((..((..(((((........)))))((((((....((....))......))))))..-.)).)))))))).....-- ( -39.10) >DroSim_CAF1 56608 113 - 1 UGCCGCUGCUUCAACUGCAGCUCCAUUUUUCCCACCAACGCCACAGAAGCCUCUUUCUUCGCUUAAUGUGCUUUGCUUCGGCUUCCCCCACAGUCG-AGCAAGUCGGUAUACUG-- ....(((((.......)))))............(((.((((.(((.((((..........))))..))))).(((((.(((((........)))))-))))))).)))......-- ( -28.70) >DroEre_CAF1 57888 114 - 1 UGCCGCUGCUUCAACUGCAGCUUCAUUUUUCCCACCGACGGCGCAGAGGCCUCUUUCUUGGCUUAAUGUGCUUUGAUUCGACUUCCCCCACAUUCC-AGCAAGUCGGUAUACUGG- ..(((((((.......)))))............((((((((((((.(((((........)))))..))))))..((.......))...........-.....)))))).....))- ( -31.90) >DroYak_CAF1 58872 100 - 1 UGCCGCUGCUUCAACUGCAGCUUCAUUUUUCCCACCGACGGCGCAGGAGCCUCUUUCUUGGCUUA---------------ACUUCUCCCACAUUAG-AGAAAGUCGGUAUACCGGU .((((((((.......)))))............(((((((((.((((((.....)))))))))..---------------..(((((........)-)))).)))))).....))) ( -32.70) >DroAna_CAF1 50684 92 - 1 UGGCGCUGCUGCAA------CUUCAUUUUUUC-----AUG-----GCAACAGCUUCCUUGGCCUAAUGUGUUUCGAUGUGGCUUCCC--AUCGUCGCAGCA--UCGCCGCCC---- .((((.((((((((------(..((((...((-----(.(-----((....)))....)))...)))).)))..(((((((....))--).))))))))))--.))))....---- ( -31.20) >consensus UGCCGCUGCUUCAACUGCAGCUUCAUUUUUCCCACCGACGCCGCAGAAGCCUCUUUCUUGGCUUAAUGUGCUUUGAUUCGGCUUCCCCCACAUUCG_AGCAAGUCGGUAUACUG__ (((((((((.......))))).........................(((((........))))).........................................))))....... (-12.54 = -13.38 + 0.84)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:57:53 2006