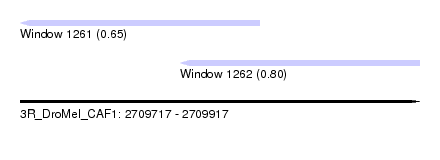
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 2,709,717 – 2,709,917 |
| Length | 200 |
| Max. P | 0.799138 |

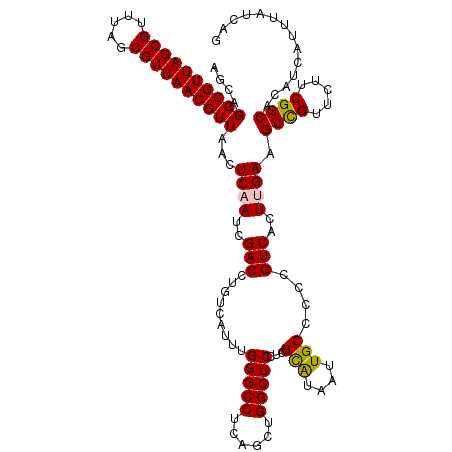
| Location | 2,709,717 – 2,709,837 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.42 |
| Mean single sequence MFE | -31.46 |
| Consensus MFE | -23.06 |
| Energy contribution | -23.06 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.651000 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 2709717 120 - 27905053 CCCGUCACUUGAAGCCGUUCUUCGGCACAUCAUUUAUCAGAGCCUUCUAUUUAUAGGCUCUCGAUCCGGCAACACUCGGCGAAACCAAGCUGAGACAGACCAAAAAAAAAAAGAGAACUU ...(((.......(((((.(....).)).......((((((((((.........))))))).)))..)))....((((((........))))))...))).................... ( -32.30) >DroSec_CAF1 20064 117 - 1 CCCGUCACUUGAAGCCGUUCUUCGGCACAUCAUUUAUCAGAGCCUUCUAUUUAUAGGAUCUCGAUCCGGCUACACUCGGCGAAACCAAGCUGACACGGCC-A--AGAAAAAGGAGAACUU ................((((((((((...((........)))))((((.......((((....))))((((....(((((........)))))...))))-.--))))...))))))).. ( -30.60) >DroSim_CAF1 21147 117 - 1 CCCGUCACUUGAAGCUGUUCUUCGGCACAUCAUUUAUCAGAGCCUUCUAUUUAUAGGAUCUCGAUCCGGCUACACUCGGCGAAACCAAGCUGACACGGCC-A--AGAAAAAGGAGAACUU ................((((((((((...((........)))))((((.......((((....))))((((....(((((........)))))...))))-.--))))...))))))).. ( -30.30) >DroEre_CAF1 21040 115 - 1 CCCGUCACUCGAAGCCGUUCUUCGGCACAUCAUUUAUCAGGGCCUUCUAUUUAUAGGCUCUCGAUCCGGCUGCACUGCACCAAACCGAGCUGGCACAGCC-C--AGAA--GGGAGAACUU ...((((((((..((((.....)))).........((((((((((.........))))))).)))..((.(((...)))))....)))).))))....((-(--....--)))....... ( -34.50) >DroYak_CAF1 20902 116 - 1 CCCGUCACUCGAAGCCGUUCUUCGGCACAUCAUUUAUCAGAGCCUUCUAUUUAUAGGCUCUCGAUCCCGCUACACUUGGCGAAACCAAGCUGGCACUGCC-A--ACAA-AAUUAGAACUU ....((....((((.....))))((((........((((((((((.........))))))).)))...((((..(((((.....))))).))))..))))-.--....-.....)).... ( -29.60) >consensus CCCGUCACUUGAAGCCGUUCUUCGGCACAUCAUUUAUCAGAGCCUUCUAUUUAUAGGCUCUCGAUCCGGCUACACUCGGCGAAACCAAGCUGACACAGCC_A__AGAAAAAGGAGAACUU ...((((((((..((((.....)))).........((((((((((.........))))))).)))....................)))).)))).......................... (-23.06 = -23.06 + 0.00)

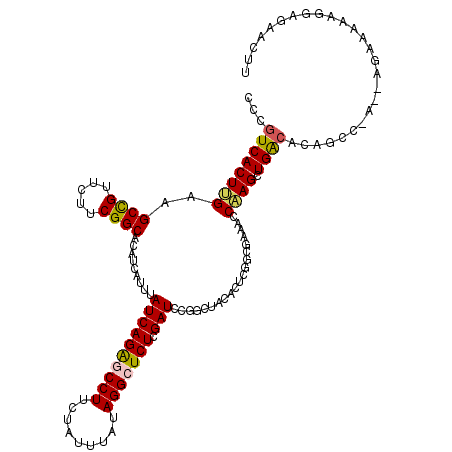
| Location | 2,709,797 – 2,709,917 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.17 |
| Mean single sequence MFE | -33.12 |
| Consensus MFE | -32.17 |
| Energy contribution | -32.08 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.10 |
| Mean z-score | -0.89 |
| Structure conservation index | 0.97 |
| SVM decision value | 0.61 |
| SVM RNA-class probability | 0.799138 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 2709797 120 - 27905053 AGCAGGCGUUAGCGUUUAGCGUUAACGUUAACUCAAUCGACCUGUCAUUUGGGCCUCAGCUGGCUCCUCGCGUAAUUGCCCCCGUCACUUGAAGCCGUUCUUCGGCACAUCAUUUAUCAG ....((((((((((.....))))))))))...((((..(((.........(((((......)))))...(((....)))....)))..)))).((((.....)))).............. ( -33.20) >DroSec_CAF1 20141 120 - 1 AGCAGGCGUUAGCGUUUAGCGUUAACGUUAACUCAAUCGACCUGUCAUUUGGGCCUCAGCUGGCUCCUCGCAUAAUUGCCCCCGUCACUUGAAGCCGUUCUUCGGCACAUCAUUUAUCAG ....((((((((((.....))))))))))...((((..(((.........(((((......)))))...(((....)))....)))..)))).((((.....)))).............. ( -33.90) >DroSim_CAF1 21224 120 - 1 AGCAGGCGUUAGCGUUUAGCGUUAACGUUAACUCAAUCGACCUGUCAUUUGGGCCUCAGCUGGCUCCUCGCAUAAUUGCCCCCGUCACUUGAAGCUGUUCUUCGGCACAUCAUUUAUCAG (((.((((((((((.....))))))))))...((((..(((.........(((((......)))))...(((....)))....)))..)))).)))((.(....).))............ ( -32.00) >DroEre_CAF1 21115 120 - 1 AGCAGGCGUUAGCGUUUAGCGUUAACGUUAACUCAAUCGACCUGUCAUUUGGGCCUCAGCUGGCUCCUCGCAUAAUUGCCCCCGUCACUCGAAGCCGUUCUUCGGCACAUCAUUUAUCAG ....((((((((((.....)))))))))).......((((..((.(....(((((......)))))...(((....)))....).)).)))).((((.....)))).............. ( -33.10) >DroYak_CAF1 20978 120 - 1 GGCAGGCGUUAGCGUUUAGCGUUAACGUUAACUCAAUCGACCUGUCAUUUGGGCCUCAGCUGGCUCCUCGUAUAAUUGCCCCCGUCACUCGAAGCCGUUCUUCGGCACAUCAUUUAUCAG ((((((((((((((.....)))))))).....((....))))))))....(((((......)))))...(.((((.((....((.....))..((((.....))))....)).))))).. ( -32.60) >DroAna_CAF1 19361 120 - 1 AGCAGGCGUUAGCGUUUAGCGUUAACGUUAACUCAAUCGACCUGUCAUUUGGGCCUCAGUUGGCUCCUCGCAUAAUUGCCCCCGUCACUUGAAGCCGUUCUUCGGCACAUCAUUUAUCAG ....((((((((((.....))))))))))...((((..(((.........(((((......)))))...(((....)))....)))..)))).((((.....)))).............. ( -33.90) >consensus AGCAGGCGUUAGCGUUUAGCGUUAACGUUAACUCAAUCGACCUGUCAUUUGGGCCUCAGCUGGCUCCUCGCAUAAUUGCCCCCGUCACUUGAAGCCGUUCUUCGGCACAUCAUUUAUCAG ....((((((((((.....))))))))))...((((..(((.........(((((......)))))...(((....)))....)))..)))).((((.....)))).............. (-32.17 = -32.08 + -0.08)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:52:14 2006