
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 21,210,073 – 21,210,200 |
| Length | 127 |
| Max. P | 0.982061 |

| Location | 21,210,073 – 21,210,166 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 63.84 |
| Mean single sequence MFE | -45.40 |
| Consensus MFE | -18.26 |
| Energy contribution | -18.14 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.55 |
| Mean z-score | -2.25 |
| Structure conservation index | 0.40 |
| SVM decision value | 1.65 |
| SVM RNA-class probability | 0.970162 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 21210073 93 + 27905053 CCCAUUCCUGUGUCUGGACCUCC---------AGGUCCGCCGGGCCCACCAGGUCCACCAGGACAGCGGGGACCUGCAGGACCCCCAGGUCCACGGGGU---------CCA ...............((((((((---------((((((.(((((....))..((((....))))..))))))))))..(((((....)))))..)))))---------)). ( -45.70) >DroVir_CAF1 50825 102 + 1 CCGAUACCAGUGCCAGGUCCACC---------AGGUCCACCGGGUCCACCGGGGCCUCCAGGACCGCGCGGACUAACCGGACCAAUGGGGCCACGAGGUAGGGAUGGUCCA .........((((..(((((...---------(((.((.((((.....)))))))))...)))))))))((((((.((..(((..((....))...)))..)).)))))). ( -44.90) >DroGri_CAF1 46635 111 + 1 CCGAUACCAGUGCCUGGUCCCCCGGGACCACCAGGACCACCUGGACCACGUGGACUGCCUGGACCACGUGGUCCAACGGGUCCAACUGGACCACGGGGCAGGGAGGGUCCA .....(((..(.((((...(((((((.(((...(((((...((((((((((((.((....)).))))))))))))...)))))...))).)).))))))))).).)))... ( -59.60) >DroWil_CAF1 38866 93 + 1 CCCAUUCCAGUUCCAGGGCCCCC---------GGGGCCACCAGGACCACCUGGCCCACCUGGUUUACGUGGCGCCACCGGACCAGUUGGCCCACGCGGA---------CAC .....(((.((.(((((((((..---------.))))).(((((....))))).....))))...))((((.((((.(......).))))))))..)))---------... ( -40.00) >DroYak_CAF1 38683 84 + 1 CCCAUUCCAGUGCCCGGUCCUCC---------UGGUCCGC---------CAGGUCCACCAGGCCAGCGGGGACCUGCAGGUCCGCCAGGUCCUCGGGGU---------CCA (((......(.(((.(((...((---------(((....)---------))))...))).))))..(((((((((((......).))))))))))))).---------... ( -41.50) >DroAna_CAF1 35573 93 + 1 CCUAUUCCAGUGCCCGGACCUCC---------GGGCCCACCCGGUCCCCCAGGGCCGCCUGGUAUGCGAGGACCUAUUGGACCCACUGGACCACGGGGC---------CAG (((..((((((((((((....))---------))))(((...(((((((((((....))))).....).)))))...)))....))))))....)))..---------... ( -40.70) >consensus CCCAUUCCAGUGCCAGGUCCUCC_________AGGUCCACCGGGACCACCAGGGCCACCAGGACAGCGGGGACCUACAGGACCAACUGGACCACGGGGU_________CCA ...........(((((((((.............(((((.((.((....)).)).((....)).......))))).............)))))...))))............ (-18.26 = -18.14 + -0.12)



| Location | 21,210,096 – 21,210,200 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 72.77 |
| Mean single sequence MFE | -53.35 |
| Consensus MFE | -26.97 |
| Energy contribution | -30.00 |
| Covariance contribution | 3.03 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.26 |
| Structure conservation index | 0.51 |
| SVM decision value | 1.84 |
| SVM RNA-class probability | 0.979551 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 21210096 104 + 27905053 AGGUCCGCCGGGCCCACCAGGUCCACCAGGACAGCGGGGACCUGCAGGACCCCCAGGUCCACGGGGU---------CCACGAGGUCCUGCCGGUGAGUCUGGCGUUAUUUCAU .....((((((((.((((((((((.((........))))))))((((((((....((.((....)).---------))....)))))))).)))).))))))))......... ( -55.10) >DroVir_CAF1 50848 110 + 1 AGGUCCACCGGGUCCACCGGGGCCUCCAGGACCGCGCGGACUAACCGGACCAAUGGGGCCACGAGGUAGGGAUGGUCCACGCGGACCACCCGGAAACGCAGGU---AUUACAG .(((((.((((.....))))((((....)).))(((.((((((.((..(((..((....))...)))..)).)))))).))))))))(((((....))..)))---....... ( -49.70) >DroSec_CAF1 36818 104 + 1 UGGCCCGCCGGGCCCACCAGGUCCACCAGGGCAGCGGGGACCUGCAGGACCACCAGGUCCACGGGGU---------CCCCGAGGUCCUGCCGGCGAGUCUGGCGUUAUUUCAG .(((.(((((((((.....)))))..((((((..(((((((((...(((((....)))))...))))---------)))))..))))))..)))).))).............. ( -59.00) >DroSim_CAF1 38454 104 + 1 UGGCCCGCCGGGCCCACCAGGUCCACCAGGGCAGCGGGGACCUGCAGGACCACCAGGUCCACGGGGU---------CCCCGAGGUCCUGCCGGCGAGUCUGGCGUUAUUUCAG .(((.(((((((((.....)))))..((((((..(((((((((...(((((....)))))...))))---------)))))..))))))..)))).))).............. ( -59.00) >DroEre_CAF1 38274 95 + 1 UGGCCCGC---------CAGGUCCACCAGGACAGCGGGGACCUGCUGGUCCGCCAGGUCCUCGGGGU---------CCACGAGGUCCUGCCGGUGAGUCUGGCAUUAUUUCAG ......((---------((((..((((.(((((((((....))))).))))(.((((.((((((...---------.).))))).)))).)))))..)))))).......... ( -51.20) >DroAna_CAF1 35596 104 + 1 GGGCCCACCCGGUCCCCCAGGGCCGCCUGGUAUGCGAGGACCUAUUGGACCCACUGGACCACGGGGC---------CAGCGAGGACCUCCAGGUGAGCCAGGUGCGUCCCCAG (((((.....)))))....((((((((((((.(((((((.((....)).(((.((((.((....)))---------))).).)).))))...))).)))))))).)))).... ( -46.10) >consensus UGGCCCGCCGGGCCCACCAGGUCCACCAGGACAGCGGGGACCUGCAGGACCACCAGGUCCACGGGGU_________CCACGAGGUCCUGCCGGUGAGUCUGGCGUUAUUUCAG .(((.((((((..((..(((((((.((........)))))))))..)).....((((.((.((.((..........)).)).)).)))))))))).))).............. (-26.97 = -30.00 + 3.03)

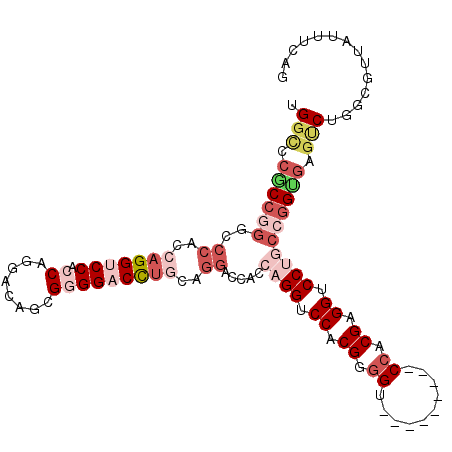

| Location | 21,210,096 – 21,210,200 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 72.77 |
| Mean single sequence MFE | -54.18 |
| Consensus MFE | -29.75 |
| Energy contribution | -31.70 |
| Covariance contribution | 1.95 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.20 |
| Structure conservation index | 0.55 |
| SVM decision value | 1.90 |
| SVM RNA-class probability | 0.982061 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 21210096 104 - 27905053 AUGAAAUAACGCCAGACUCACCGGCAGGACCUCGUGG---------ACCCCGUGGACCUGGGGGUCCUGCAGGUCCCCGCUGUCCUGGUGGACCUGGUGGGCCCGGCGGACCU .........((((.(.((((((.((((((((((..((---------.((....)).))..))))))))))(((((((((......))).)))))))))))).).))))..... ( -55.10) >DroVir_CAF1 50848 110 - 1 CUGUAAU---ACCUGCGUUUCCGGGUGGUCCGCGUGGACCAUCCCUACCUCGUGGCCCCAUUGGUCCGGUUAGUCCGCGCGGUCCUGGAGGCCCCGGUGGACCCGGUGGACCU ..(((..---...)))...((((((....((((((((((...((..(((..(((....))).)))..))...)))))))))).))))))((((((((.....)))).)).)). ( -49.90) >DroSec_CAF1 36818 104 - 1 CUGAAAUAACGCCAGACUCGCCGGCAGGACCUCGGGG---------ACCCCGUGGACCUGGUGGUCCUGCAGGUCCCCGCUGCCCUGGUGGACCUGGUGGGCCCGGCGGGCCA .........((((((..((((((((((.....(((((---------(((..(((((((....))))).)).)))))))))))))..)))))..))))))(((((...))))). ( -59.80) >DroSim_CAF1 38454 104 - 1 CUGAAAUAACGCCAGACUCGCCGGCAGGACCUCGGGG---------ACCCCGUGGACCUGGUGGUCCUGCAGGUCCCCGCUGCCCUGGUGGACCUGGUGGGCCCGGCGGGCCA .........((((((..((((((((((.....(((((---------(((..(((((((....))))).)).)))))))))))))..)))))..))))))(((((...))))). ( -59.80) >DroEre_CAF1 38274 95 - 1 CUGAAAUAAUGCCAGACUCACCGGCAGGACCUCGUGG---------ACCCCGAGGACCUGGCGGACCAGCAGGUCCCCGCUGUCCUGGUGGACCUG---------GCGGGCCA .........((((((..((((((.((((.(((((.(.---------...)))))).)))).)((((.(((.(....).))))))).)))))..)))---------)))..... ( -45.30) >DroAna_CAF1 35596 104 - 1 CUGGGGACGCACCUGGCUCACCUGGAGGUCCUCGCUG---------GCCCCGUGGUCCAGUGGGUCCAAUAGGUCCUCGCAUACCAGGCGGCCCUGGGGGACCGGGUGGGCCC ..(((......)))(((((((((...(((((((.(.(---------(((((.((((...(((((.((....)).)).)))..)))))).))))..))))))))))))))))). ( -55.20) >consensus CUGAAAUAACGCCAGACUCACCGGCAGGACCUCGUGG_________ACCCCGUGGACCUGGUGGUCCUGCAGGUCCCCGCUGUCCUGGUGGACCUGGUGGGCCCGGCGGGCCA .........((((...........((((.((.((.((..........)).)).)).))))..(((((..((((((((((......))).)))))))..))))).))))..... (-29.75 = -31.70 + 1.95)


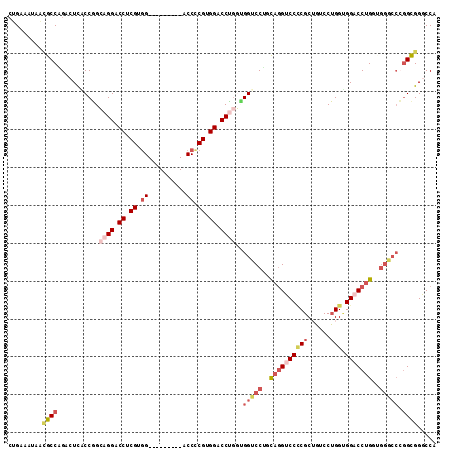
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:57:39 2006