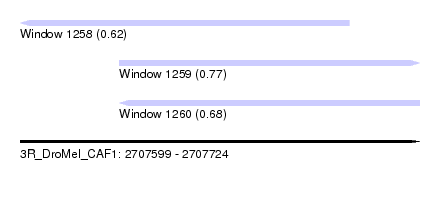
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 2,707,599 – 2,707,724 |
| Length | 125 |
| Max. P | 0.765523 |

| Location | 2,707,599 – 2,707,702 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 85.59 |
| Mean single sequence MFE | -29.26 |
| Consensus MFE | -21.54 |
| Energy contribution | -21.98 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.617650 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 2707599 103 - 27905053 --CACCAUGAUGCC-AUGCCAAUGGCCGCCAUACGCACCACUGUGAGUGCGUGAGUAUCCGCAGAAGCCGUGUGCACCGAAAAAAAAUAUAUCGUAAGCAUGAAUA--------- --...((((..(((-((....))))).((.(((((.....(((((..(((....)))..)))))....))))))).......................))))....--------- ( -29.50) >DroSec_CAF1 17984 113 - 1 ACCACCAUGAUGCC-AUGCCAAUGGCCGCCAUACGCACCAUUGUGAGUGCGUGAGUAUCCGCAGAAGCCGUGUGCACAGAAAAC-AAUUUUUCUAAAUCAUGAAUAUUUAAAUUA .....(((((((((-((....))))).((.(((((.....(((((..(((....)))..)))))....)))))))..((((((.-...))))))..))))))............. ( -31.90) >DroSim_CAF1 19063 113 - 1 ACCACCAUGAUGCC-AUGCCAAUGGCCGCCAUACGCACCAUUGUGAGUGCGUGAGUAUCCGCAGAAGCCGUGUGCACCGAAAAC-AAUUUUUCUAAAUCAUGAAUAUUUAAAUUA .....(((((((((-((....))))).((.(((((.....(((((..(((....)))..)))))....)))))))...(((((.-...)))))...))))))............. ( -30.80) >DroEre_CAF1 18863 107 - 1 --CACCAUGAUGCCCAUGCCAAUGGCCGCCAUACGCACAAUUGUGAGUGCGUGAGUAUCCGCAGAAGCCGUGUGCACCGGAAAA-AAUAUUUCGAAGGCAUGACUAUUUA----- --......((((..((((((.((((...))))..(((((.(((((..(((....)))..)))))......)))))..(((((..-....)))))..))))))..))))..----- ( -31.40) >DroYak_CAF1 18731 109 - 1 ACCACCAUGAUGCCAAUGCCAAUGGCCGCCAUACGCACAAUUGUGUGUGCGUGAGUAUCUGCAGAAGCCGUGACACCGGGAAAA-AAUAUUUCGAAAACAUAAAUAUUUA----- .....((((..((...(((.....(((((((((((((....)))))))).))).))....)))...))))))....((..(...-....)..))................----- ( -22.70) >consensus ACCACCAUGAUGCC_AUGCCAAUGGCCGCCAUACGCACCAUUGUGAGUGCGUGAGUAUCCGCAGAAGCCGUGUGCACCGAAAAA_AAUAUUUCGAAAGCAUGAAUAUUUA_____ .....((((.......(((..(((((.((.....))....(((((..(((....)))..)))))..)))))..)))..((((.......)))).....))))............. (-21.54 = -21.98 + 0.44)

| Location | 2,707,630 – 2,707,724 |
|---|---|
| Length | 94 |
| Sequences | 4 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 88.75 |
| Mean single sequence MFE | -43.60 |
| Consensus MFE | -32.96 |
| Energy contribution | -33.40 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.51 |
| SVM RNA-class probability | 0.765523 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 2707630 94 + 27905053 GCACACGGCUUCUGCGGAUACUCACGCACUCACAGUGGUGCGUAUGGCGGCCAUUGGCAU-GGCAUCAUGGUG-----------GUGCCAUGAUGGUGGUGC---CGUU ....(((((...(((.(.(((.(((.(((.....)))))).)))).)))((((((..(((-((((((.....)-----------))))))))..))))))))---))). ( -41.90) >DroSec_CAF1 18023 105 + 1 GCACACGGCUUCUGCGGAUACUCACGCACUCACAAUGGUGCGUAUGGCGGCCAUUGGCAU-GGCAUCAUGGUGGUGCUCCGGUGGUGCCAUGGUGGUGGUGC---CGUU .....(.((....)).)......(((((((......)))))))((((((.((((((.(((-((((((((..(((....)))))))))))))).)))))))))---))). ( -47.00) >DroSim_CAF1 19102 108 + 1 GCACACGGCUUCUGCGGAUACUCACGCACUCACAAUGGUGCGUAUGGCGGCCAUUGGCAU-GGCAUCAUGGUGGUGCUCCGGUGGUGCCAUGGUGGUGGUGCCGCCGUU ....(((((..............(((((((......)))))))..((((.((((((.(((-((((((((..(((....)))))))))))))).))))))))))))))). ( -51.40) >DroEre_CAF1 18897 95 + 1 GCACACGGCUUCUGCGGAUACUCACGCACUCACAAUUGUGCGUAUGGCGGCCAUUGGCAUGGGCAUCAUGGUG-----------GUGGCAUGGUGGUGGUGC---UGUU ((((((.(((..(((.....(..((((((........))))))..)((.(((((..((....))...))))).-----------)).))).))).)).))))---.... ( -34.10) >consensus GCACACGGCUUCUGCGGAUACUCACGCACUCACAAUGGUGCGUAUGGCGGCCAUUGGCAU_GGCAUCAUGGUG___________GUGCCAUGGUGGUGGUGC___CGUU ((((((.(((...((.(...(..((((((........))))))..).).))....))).....((((((((((.(((....))).)))))))))))).))))....... (-32.96 = -33.40 + 0.44)

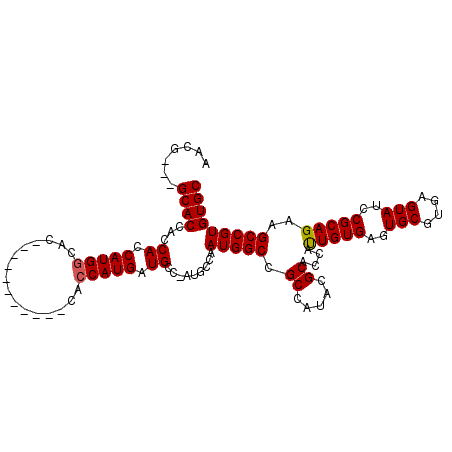
| Location | 2,707,630 – 2,707,724 |
|---|---|
| Length | 94 |
| Sequences | 4 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 88.75 |
| Mean single sequence MFE | -34.71 |
| Consensus MFE | -25.75 |
| Energy contribution | -25.82 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.31 |
| SVM RNA-class probability | 0.684665 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 2707630 94 - 27905053 AACG---GCACCACCAUCAUGGCAC-----------CACCAUGAUGCC-AUGCCAAUGGCCGCCAUACGCACCACUGUGAGUGCGUGAGUAUCCGCAGAAGCCGUGUGC ...(---((......(((((((...-----------..)))))))(((-((....))))).)))...(((((..(((((..(((....)))..))))).....))))). ( -37.90) >DroSec_CAF1 18023 105 - 1 AACG---GCACCACCACCAUGGCACCACCGGAGCACCACCAUGAUGCC-AUGCCAAUGGCCGCCAUACGCACCAUUGUGAGUGCGUGAGUAUCCGCAGAAGCCGUGUGC ...(---((.(((....(((((((.((..((.......)).)).))))-)))....)))..)))...(((((..(((((..(((....)))..))))).....))))). ( -35.50) >DroSim_CAF1 19102 108 - 1 AACGGCGGCACCACCACCAUGGCACCACCGGAGCACCACCAUGAUGCC-AUGCCAAUGGCCGCCAUACGCACCAUUGUGAGUGCGUGAGUAUCCGCAGAAGCCGUGUGC ...((((((........(((((((.((..((.......)).)).))))-)))......))))))...(((((..(((((..(((....)))..))))).....))))). ( -41.14) >DroEre_CAF1 18897 95 - 1 AACA---GCACCACCACCAUGCCAC-----------CACCAUGAUGCCCAUGCCAAUGGCCGCCAUACGCACAAUUGUGAGUGCGUGAGUAUCCGCAGAAGCCGUGUGC ....---.............((((.-----------...((((.....))))....))))...((((((.....(((((..(((....)))..)))))....)))))). ( -24.30) >consensus AACG___GCACCACCACCAUGGCAC___________CACCAUGAUGCC_AUGCCAAUGGCCGCCAUACGCACCAUUGUGAGUGCGUGAGUAUCCGCAGAAGCCGUGUGC .......((((...((.(((((................))))).)).........(((((.((.....))....(((((..(((....)))..)))))..))))))))) (-25.75 = -25.82 + 0.06)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:52:12 2006