| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 21,118,503 – 21,118,616 |
| Length | 113 |
| Max. P | 0.735053 |

| Location | 21,118,503 – 21,118,616 |
|---|---|
| Length | 113 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.73 |
| Mean single sequence MFE | -33.24 |
| Consensus MFE | -18.88 |
| Energy contribution | -20.10 |
| Covariance contribution | 1.22 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.49 |
| Structure conservation index | 0.57 |
| SVM decision value | 0.43 |
| SVM RNA-class probability | 0.735053 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
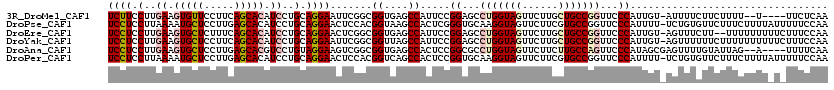
>3R_DroMel_CAF1 21118503 113 + 27905053 UCUUCCUUGAAGUGUUCCUUCAGCACAUCCUGCAGGAAUUCGGCGGUGAGCCAUUCCGGAGCCUGGUAGUUCUUGCUGCCGGUUCCCAUUGU-AUUUUCUUCUUUU--U----UUCUCAA ........((((.((((((.(((......))).))))))..(((.....))).....((((((.((((((....))))))))))))......-.....))))....--.----....... ( -34.30) >DroPse_CAF1 17893 119 + 1 UCCUCCUUAAAAUGCUCCUUGAGCACAUCCUGCAGGAACUCCACGGUAAGCCACUCGGGUGCAAGGUAGUUCUUCGUGCCGGUUCCCAUUUU-UCUGUGUUCUUUCUUUUAUUUUUCCAA ........((((((......(((((((.......((((((....((....)).....((..(((((....)))).)..))))))))......-..))))))).......))))))..... ( -25.38) >DroEre_CAF1 26206 117 + 1 UCCUCCUUGAAGUGCUCUUUCAGCACAUCCUGCAGGAACUCGGCGGUGAGCCAUUCCGGAGCCUGGUAGUUCUUGCUGCCGGUUCCCAUUGU-AGUUUCUU--UUUUUUUUUCUUUCCAA ........((((((((.....)))))...(((((((((...(((.....))).))))((((((.((((((....))))))))))))...)))-)).)))..--................. ( -37.90) >DroYak_CAF1 27583 119 + 1 UCCUCCUUGAAGUGCUCCUUCAGCACAUCCUGCAGGAAUUCGGCGGUUAGCCAUUCCGGAGCCUGGUAGUUCUUGCUGCCGGUUCCCAUUGU-AGUUUUUUCUUUUUUUUUUCUUUCCAA ........((((((((.....)))))...((((((((((..(((.....))))))))((((((.((((((....))))))))))))...)))-))....))).................. ( -39.10) >DroAna_CAF1 26641 114 + 1 UCCUCCUUGAAGUGCUCCUUGAGCACGUCCUGUAGGAAGUCGGCGGUGAGCCACUCCGGCGCCUGGUAGUUCUUCUUGCCAGUUCCCAUAGCGAGUUUUGUAUUAG--A----UUUUCAA .......((((((((((...)))))).((((((.(((((((((.((((...)))))))))..(((((((......))))))))))).)))).))............--.----..)))). ( -35.90) >DroPer_CAF1 18379 119 + 1 UCCUCCUUAAAAUGCUCCUUGAGCACAUCCUGCAGGAACUCCACGGUCAGCCACUCCGGUGCAAGGUAGUUCUUCGUGCCGGUUCCCAUUUU-UCUGUGUUCUUUCUUUUAUUUUUCCAA ........((((((......(((((((.......(((((.....((....))....(((..(((((....)))).)..))))))))......-..))))))).......))))))..... ( -26.88) >consensus UCCUCCUUGAAGUGCUCCUUCAGCACAUCCUGCAGGAACUCGGCGGUGAGCCACUCCGGAGCCUGGUAGUUCUUCCUGCCGGUUCCCAUUGU_AGUUUCUUCUUUUUUUU_U_UUUCCAA ((((.(..((.(((((.....))))).))..).)))).......((....)).....((...(((((((......)))))))...))................................. (-18.88 = -20.10 + 1.22)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:57:09 2006