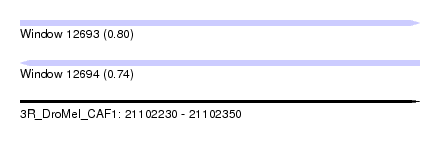
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 21,102,230 – 21,102,350 |
| Length | 120 |
| Max. P | 0.800576 |

| Location | 21,102,230 – 21,102,350 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.50 |
| Mean single sequence MFE | -45.42 |
| Consensus MFE | -30.49 |
| Energy contribution | -30.92 |
| Covariance contribution | 0.42 |
| Combinations/Pair | 1.32 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.62 |
| SVM RNA-class probability | 0.800576 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
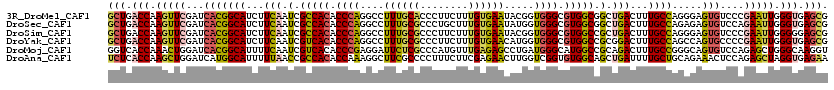
>3R_DroMel_CAF1 21102230 120 + 27905053 CGCUCACCCAAUUCGGGACACUCCCUGGCAAAGUCAGCCGCCACGCCCACCGUAUUCACAAAGAAGGGUGCAAAGGCCUGGGUGUGGCGAUUGAAGAUGCCGUGAUCGAACUUGGUCAGC .(((.(((...((((((.....))).((((...(((..(((((((((((((((((((........))))))...))..)))))))))))..)))...))))......)))...))).))) ( -50.10) >DroSec_CAF1 11264 120 + 1 CGCUCACCCAAUUCUGGACACUCUCUGGCAAAGUCAGCCGCCACGCCCACCAUAUUCACAAAGCAGGGCGCAAAGGCCUGGGUGUGGCGAUUGAAGAUGCCGUGAUCGAACUUGGUCAGC .(((.(((...(((.((.(((.....((((...(((..(((((((((((.........(......)(((......))))))))))))))..)))...))))))).)))))...))).))) ( -43.60) >DroSim_CAF1 11697 120 + 1 CGCUCCCCCAAUUCGGGACACUCCCUGGCAAAGUCAGCGGCCACGCCCACCGUAUUCACAAAGAAGGGCGCAAAGGCCUGGGUGUGGCGAUUGAAGAUGCCGUGAUCGAACUUGGUCAGC .(((...((((((((((.....))).((((...((((..(((((((((......(((.....)))((((......)))))))))))))..))))...))))......))).))))..))) ( -47.40) >DroYak_CAF1 11725 120 + 1 CGCUCACCCAAUUCGGGGCACUGGCUGGCAAAGUCCGCGGCCACGCCCACCAUGUUCACAAAGAAGGGCGCAAAGGCCUGGGUGUGACGAUUGAAGAUGCCGUGAUCGAACUUGGUCAGC .((...(((.....)))))....((((((.((((((((((((((((((......(((.....)))((((......))))))))))).(.......)..))))))...).)))).)))))) ( -41.40) >DroMoj_CAF1 5253 120 + 1 ACCUUGCCCAGCUCUGGACACUGCCCGGCAAAGUCUGCGGCCAUGCCCAUCAGGCUCUCAAACAUGGGCGAGAAUCCUCGGGUGUGACGAUUGAAAAUGCCGUGAUCCAGUUUGGUGACC .......((((..((((((((.....((((...((..((..(((((((.....((((........))))(((....)))))))))).))...))...))))))).))))).))))..... ( -40.80) >DroAna_CAF1 9549 120 + 1 UUCUCACCUAGCUCUGGAGUUUCUGCAGCAAAAUCAGCUGCCACACCGACCAAGUUCUCGAAGAAAGGGGCGAAGCCUUUGGUGUGGCGGUUAAAAAUGCCAUGAUCCAGCUUGGUGAGA .(((((((.((((..(((......(((........((((((((((((((((...(((.....))).))(((...))).)))))))))))))).....))).....))))))).))))))) ( -49.22) >consensus CGCUCACCCAAUUCGGGACACUCCCUGGCAAAGUCAGCGGCCACGCCCACCAUAUUCACAAAGAAGGGCGCAAAGGCCUGGGUGUGGCGAUUGAAGAUGCCGUGAUCGAACUUGGUCAGC .(((.(((...(((.((.(((.....((((...((((..(((((((((......(((.....)))((((......)))))))))))))..))))...))))))).)))))...))).))) (-30.49 = -30.92 + 0.42)



| Location | 21,102,230 – 21,102,350 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.50 |
| Mean single sequence MFE | -46.90 |
| Consensus MFE | -31.87 |
| Energy contribution | -31.85 |
| Covariance contribution | -0.02 |
| Combinations/Pair | 1.35 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.45 |
| SVM RNA-class probability | 0.740195 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
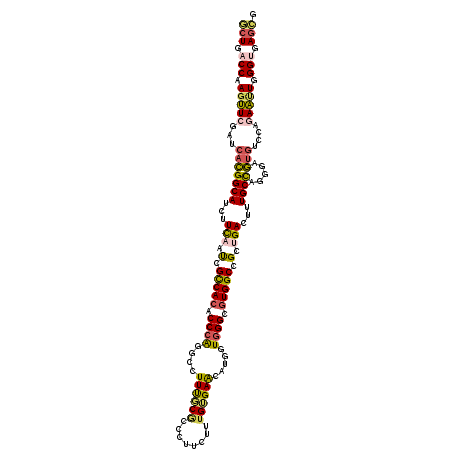
>3R_DroMel_CAF1 21102230 120 - 27905053 GCUGACCAAGUUCGAUCACGGCAUCUUCAAUCGCCACACCCAGGCCUUUGCACCCUUCUUUGUGAAUACGGUGGGCGUGGCGGCUGACUUUGCCAGGGAGUGUCCCGAAUUGGGUGAGCG (((.(((.(((((......((((...(((..((((((.(((..(((((..((........))..))...)))))).))))))..)))...)))).(((.....)))))))).))).))). ( -53.80) >DroSec_CAF1 11264 120 - 1 GCUGACCAAGUUCGAUCACGGCAUCUUCAAUCGCCACACCCAGGCCUUUGCGCCCUGCUUUGUGAAUAUGGUGGGCGUGGCGGCUGACUUUGCCAGAGAGUGUCCAGAAUUGGGUGAGCG (((.(((.(((((...(((((((...(((..((((((.(((..(((....(((........))).....)))))).))))))..)))...)))).....)))....))))).))).))). ( -49.00) >DroSim_CAF1 11697 120 - 1 GCUGACCAAGUUCGAUCACGGCAUCUUCAAUCGCCACACCCAGGCCUUUGCGCCCUUCUUUGUGAAUACGGUGGGCGUGGCCGCUGACUUUGCCAGGGAGUGUCCCGAAUUGGGGGAGCG (((..((.((((((..((((((..........)))...(((.((((...((((((....((((....)))).))))))))))((.......))..))).)))...)))))).))..))). ( -46.60) >DroYak_CAF1 11725 120 - 1 GCUGACCAAGUUCGAUCACGGCAUCUUCAAUCGUCACACCCAGGCCUUUGCGCCCUUCUUUGUGAACAUGGUGGGCGUGGCCGCGGACUUUGCCAGCCAGUGCCCCGAAUUGGGUGAGCG (((.(((.((((((..((((((....................((((...((((((..((.((....)).)).))))))))))..((......)).))).)))...)))))).))).))). ( -47.70) >DroMoj_CAF1 5253 120 - 1 GGUCACCAAACUGGAUCACGGCAUUUUCAAUCGUCACACCCGAGGAUUCUCGCCCAUGUUUGAGAGCCUGAUGGGCAUGGCCGCAGACUUUGCCGGGCAGUGUCCAGAGCUGGGCAAGGU (((((.((((((((.....(((..........))).....((((....)))).))).)))))...(((.....))).)))))....((((((((.(((..........))).)))))))) ( -39.80) >DroAna_CAF1 9549 120 - 1 UCUCACCAAGCUGGAUCAUGGCAUUUUUAACCGCCACACCAAAGGCUUCGCCCCUUUCUUCGAGAACUUGGUCGGUGUGGCAGCUGAUUUUGCUGCAGAAACUCCAGAGCUAGGUGAGAA (((((((.(((((((.....((..........((((((((...(((...)))((.(((.....)))...))..))))))))(((.......)))))......)))..)))).))))))). ( -44.50) >consensus GCUGACCAAGUUCGAUCACGGCAUCUUCAAUCGCCACACCCAGGCCUUUGCGCCCUUCUUUGUGAACAUGGUGGGCGUGGCCGCUGACUUUGCCAGGGAGUGUCCAGAAUUGGGUGAGCG (((.(((.(((((...(((((((...(((.(.(((((.((((....((((((........)))))).....)))).))))).).)))...)))).....)))....))))).))).))). (-31.87 = -31.85 + -0.02)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:57:03 2006