
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 2,704,532 – 2,704,652 |
| Length | 120 |
| Max. P | 0.500000 |

| Location | 2,704,532 – 2,704,652 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.89 |
| Mean single sequence MFE | -29.75 |
| Consensus MFE | -27.47 |
| Energy contribution | -27.22 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.09 |
| Mean z-score | -0.92 |
| Structure conservation index | 0.92 |
| SVM decision value | -0.07 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 2704532 120 + 27905053 UGCUUUUGAUGAUGAUGGCAUUUCCUUUGGUAGUUGCGUACGUCAUAGAUUAAUGGCAGCAAUCCAUCAAAAUCACUCGAUGGACACGACGAACAGCCGAAAAUUCGGAAGAAGCAGCCA ...............((((......((((((.(((.(((.((((((......))))).....((((((..........))))))...))))))).))))))..(((....)))...)))) ( -31.30) >DroPse_CAF1 43967 120 + 1 UGCUUUUGAUGAUGAUGGCAUUUCCUUUGGUAGUUGCGUACGUCAUAGAUUAAUGACAGCAAUUCAUCAAAAUCACUCGAUGGACAUGACGAACAGCCGAAAAUUCAGGAGAAGAAACUG ..(((((((......((((...(((((((((((((((....(((((......))))).)))))).))))))(((....))))))..((.....)))))).....)))))))......... ( -26.30) >DroEre_CAF1 15396 120 + 1 UGCUUUUGAUGAUGAUGGCAUUUCCUUUGGUAGUUGCGUACGUCAUAGAUUAAUGGCAGCAAUCCAUCAAAAUCACUCGAUGGACACGAAGAACAGCCGAAAAUUCGGAAGAAGCAGCAA (((((((..(((...((((..(((.((((((.(((((....(((((......))))).)))))(((((..........))))))).)))))))..)))).....)))..))))))).... ( -31.60) >DroYak_CAF1 15573 120 + 1 UGCUUUUGAUGAUGAUGGCAUUUCCUUUGGUAGUUGCGUACGUCAUAGAUUAAUGGCAGCAAUCCAUCAAAAUCACUCGAUGGACACGACGAACAGCCGAAAAUUCGGAAGAAGCAGCAA (((((((..(((....((.....))((((((.(((.(((.((((((......))))).....((((((..........))))))...))))))).))))))...)))..))))))).... ( -31.00) >DroAna_CAF1 14825 120 + 1 UGCUUUUGAUGAUGAUGGCAUUUCCUUUGGUAGUUGCGUACGUCAUAGAUUAAUGACAGCAAUUCAUCAAAAUCACUCGAUGGACAUGACGAACAGCCGAAAAUUCAGCAGAAGCAGUCA ((((((((.(((...((((...(((((((((((((((....(((((......))))).)))))).))))))(((....))))))..((.....)))))).....))).)))))))).... ( -32.00) >DroPer_CAF1 43932 120 + 1 UGCUUUUGAUGAUGAUGGCAUUUCCUUUGGUAGUUGCGUACGUCAUAGAUUAAUGACAGCAAUUCAUCAAAAUCACUCGAUGGACAUGACGAACAGCCGAAAAUUCAGGAGAAGAAACCG ..(((((((......((((...(((((((((((((((....(((((......))))).)))))).))))))(((....))))))..((.....)))))).....)))))))......... ( -26.30) >consensus UGCUUUUGAUGAUGAUGGCAUUUCCUUUGGUAGUUGCGUACGUCAUAGAUUAAUGACAGCAAUCCAUCAAAAUCACUCGAUGGACACGACGAACAGCCGAAAAUUCAGAAGAAGCAGCCA (((((((..(((....((.....))((((((.(((.(((..(((((......))))).....((((((..........))))))....)))))).))))))...)))..))))))).... (-27.47 = -27.22 + -0.25)

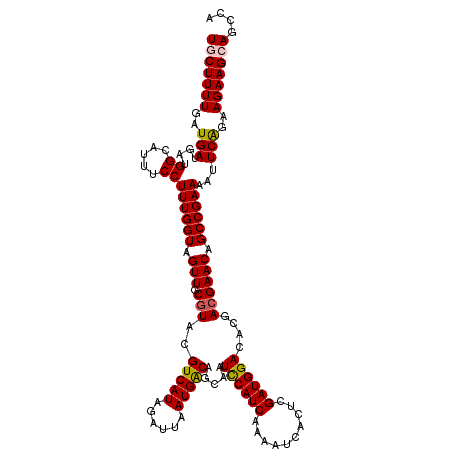
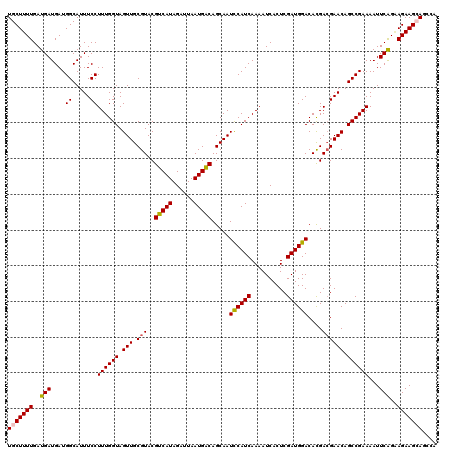
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:52:08 2006