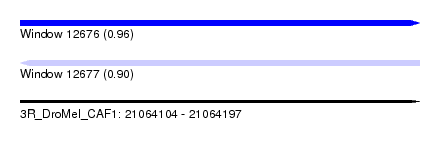
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 21,064,104 – 21,064,197 |
| Length | 93 |
| Max. P | 0.959269 |

| Location | 21,064,104 – 21,064,197 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 77.61 |
| Mean single sequence MFE | -34.23 |
| Consensus MFE | -21.93 |
| Energy contribution | -22.60 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.19 |
| Structure conservation index | 0.64 |
| SVM decision value | 1.50 |
| SVM RNA-class probability | 0.959269 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 21064104 93 + 27905053 GCAGCGCAAUGGACGCACCUACCUGAGCGUGGAUAAGCUCAAGGUUCUGGUGGAGCCACAAAAGUAGGUGC--AGUUCGGA---U------U---------UUAGCUGAGAUA .((((....(((((((((((((.(((((........))))).(((((.....)))))......))))))))--.)))))..---.------.---------...))))..... ( -38.20) >DroVir_CAF1 18138 93 + 1 GUCGCGCAACGGACGCACCUAUCUGAGCGUGGACAAGCUCAAGGUGCUCGUGGAGCCACAGAAGUAAGUUG--AGAGCGGA---U------C---------AAAGGUUAUACA .((((.((((..((((((((...(((((........)))))))))))..(((....)))....))..))))--...)))).---.------.---------............ ( -32.60) >DroSec_CAF1 17460 93 + 1 GCAGCGCAAUGGACGCACCUACCUGAGCGUGGAUAAGCUCAAGGUGCUGGUGGAGCCACAAAAGUGGGUGC--AGUUCGGA---U------U---------UUAGCUGACUCA .((((....(((((((((((((((((((........)))).))))(((.(((....)))...)))))))))--.)))))..---.------.---------...))))..... ( -36.40) >DroSim_CAF1 16752 93 + 1 GCAGCGCAAUGGACGCACCUAUUUGAGCGUGGAUAAGCUCAAGGUGCUGGUGGAGCCACAAAAGUGGGUGC--AGUUCGGA---U------U---------UUAGCUGACUCA .((((....(((((((((((...(((((........)))))))))))...((.(.((((....)))).).)--))))))..---.------.---------...))))..... ( -35.10) >DroEre_CAF1 18090 103 + 1 GGUGCGCAAUGGACGCACCUACCUGAGCGUGGAUAAGCUCAAGGUGCUGGUGGAGCCACAAAAGUAAGUGC--AGCUCAGA---UUGGUGUCCUUUCAUGUUAAGCUG----- (((..(((..((((((((((((((((((........)))).)))))..))).(((((((........))).--.))))...---...)))))).....)))...))).----- ( -33.10) >DroAna_CAF1 17359 95 + 1 GCAGCGCAACGGUCGCACCUUCCUGAGCGUGGACAAGCUCAAGGUGCUUGUGGAGCCCCAAAAGUAAGUGCCAAGCUGGGGGAGA------U---------UUAGAGAA---A ...((((....).)))..((((((.(((.(((((((((.......))))))((....))...........))).))).)))))).------.---------........---. ( -30.00) >consensus GCAGCGCAAUGGACGCACCUACCUGAGCGUGGAUAAGCUCAAGGUGCUGGUGGAGCCACAAAAGUAAGUGC__AGUUCGGA___U______U_________UUAGCUGA__CA ...((((.......((((((...(((((........)))))))))))..(((....)))........)))).......................................... (-21.93 = -22.60 + 0.67)

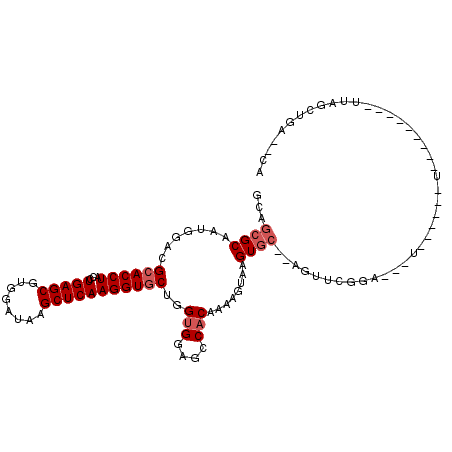
| Location | 21,064,104 – 21,064,197 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 77.61 |
| Mean single sequence MFE | -27.85 |
| Consensus MFE | -18.54 |
| Energy contribution | -18.73 |
| Covariance contribution | 0.19 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.67 |
| SVM decision value | 1.00 |
| SVM RNA-class probability | 0.897813 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 21064104 93 - 27905053 UAUCUCAGCUAA---------A------A---UCCGAACU--GCACCUACUUUUGUGGCUCCACCAGAACCUUGAGCUUAUCCACGCUCAGGUAGGUGCGUCCAUUGCGCUGC .....((((...---------.------.---........--(((((((((((.(((....))).)))...((((((........))))))))))))))((.....)))))). ( -28.40) >DroVir_CAF1 18138 93 - 1 UGUAUAACCUUU---------G------A---UCCGCUCU--CAACUUACUUCUGUGGCUCCACGAGCACCUUGAGCUUGUCCACGCUCAGAUAGGUGCGUCCGUUGCGCGAC ............---------.------.---..(((...--((((..((...((((....)))).(((((((((((........)))))...))))))))..)))).))).. ( -27.00) >DroSec_CAF1 17460 93 - 1 UGAGUCAGCUAA---------A------A---UCCGAACU--GCACCCACUUUUGUGGCUCCACCAGCACCUUGAGCUUAUCCACGCUCAGGUAGGUGCGUCCAUUGCGCUGC .....((((...---------.------.---........--((((((((....)))(((.....)))((((.((((........)))))))).)))))((.....)))))). ( -26.70) >DroSim_CAF1 16752 93 - 1 UGAGUCAGCUAA---------A------A---UCCGAACU--GCACCCACUUUUGUGGCUCCACCAGCACCUUGAGCUUAUCCACGCUCAAAUAGGUGCGUCCAUUGCGCUGC .....((((...---------.------.---........--....((((....))))........(((((((((((........)))))...)))))).........)))). ( -26.30) >DroEre_CAF1 18090 103 - 1 -----CAGCUUAACAUGAAAGGACACCAA---UCUGAGCU--GCACUUACUUUUGUGGCUCCACCAGCACCUUGAGCUUAUCCACGCUCAGGUAGGUGCGUCCAUUGCGCACC -----...............((((.....---...((((.--.((........))..)))).....(((((((((((........)))))...)))))))))).......... ( -33.60) >DroAna_CAF1 17359 95 - 1 U---UUCUCUAA---------A------UCUCCCCCAGCUUGGCACUUACUUUUGGGGCUCCACAAGCACCUUGAGCUUGUCCACGCUCAGGAAGGUGCGACCGUUGCGCUGC .---........---------.------.......((((.((((.........(((....)))...(((((((((((........)))))...))))))....)))).)))). ( -25.10) >consensus UG__UCAGCUAA_________A______A___UCCGAACU__GCACCUACUUUUGUGGCUCCACCAGCACCUUGAGCUUAUCCACGCUCAGGUAGGUGCGUCCAUUGCGCUGC ..........................................(((.((((....))))........(((((((((((........)))))...))))))......)))..... (-18.54 = -18.73 + 0.19)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:56:45 2006