
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 21,057,172 – 21,057,308 |
| Length | 136 |
| Max. P | 0.989265 |

| Location | 21,057,172 – 21,057,282 |
|---|---|
| Length | 110 |
| Sequences | 4 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 88.54 |
| Mean single sequence MFE | -33.50 |
| Consensus MFE | -27.29 |
| Energy contribution | -27.85 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.09 |
| Mean z-score | -3.22 |
| Structure conservation index | 0.81 |
| SVM decision value | 2.16 |
| SVM RNA-class probability | 0.989265 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 21057172 110 + 27905053 UCAUCCAUAUAA-AAUAGGUAUGAAGAUCAGUCUGUUAUACCCUCAUGGACUUAAAGAGUAUAUAGACAACAGCAGCCACUGAAGACGGUGGCAAUGUUUUUGUUU---UUGUU ...(((((....-....((((((((((....))).)))))))...)))))..........(((.((((((.(((((((((((....)))))))..)))).))))))---.))). ( -28.20) >DroSec_CAF1 10544 111 + 1 UCAUCCAUAUGAAAAUAGGUAUGCAGAUCAGUCUGUUAUACCCUCAUGGACUUAAAGAGUAUAUAGACAACAGCAGCCACUGAAGACGGUGGCAAUGUUUUUGUUG---UUGUU ...(((((((....)))((((((((((....)))).))))))....))))...............(((((((((((((((((....)))))))........)))))---))))) ( -35.00) >DroSim_CAF1 9751 110 + 1 UCAUCCAUAUGA-AAUAGGUAUGCAGAUCAGUCUGUUAUACCCUCAUGGACUUAAAGAGUAUAUAGACAACAGCAGCCACUGAAGACGGUGGCAAUGUUUUUGUUG---UUGUU ...((((..(((-....((((((((((....)))).)))))).)))))))...............(((((((((((((((((....)))))))........)))))---))))) ( -34.60) >DroYak_CAF1 10064 108 + 1 UCUUCCAUAUAA-AAUGGGUAUGCAGAUCGGUCUAGCAUACCUCUAUGGAUUCAAAGAGU-----CACAACAGCAGCCACUGAAGACGGUGGCAAUGCUUUUGUUUUUGUUGUU ...((((((...-...(((((((((((....))).)))))))).))))))....((.((.-----.((((.(((((((((((....)))))))..)))).))))..)).))... ( -36.20) >consensus UCAUCCAUAUAA_AAUAGGUAUGCAGAUCAGUCUGUUAUACCCUCAUGGACUUAAAGAGUAUAUAGACAACAGCAGCCACUGAAGACGGUGGCAAUGUUUUUGUUG___UUGUU ...(((((((....)))((((((((((....)))).))))))....))))...............(((((.(((((((((((....)))))))..)))).)))))......... (-27.29 = -27.85 + 0.56)

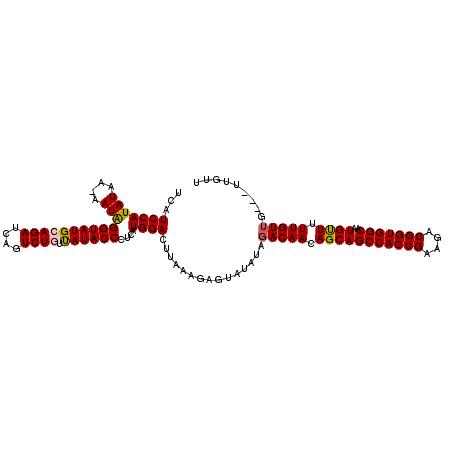
| Location | 21,057,172 – 21,057,282 |
|---|---|
| Length | 110 |
| Sequences | 4 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 88.54 |
| Mean single sequence MFE | -27.00 |
| Consensus MFE | -19.92 |
| Energy contribution | -20.18 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.29 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.87 |
| SVM RNA-class probability | 0.871750 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 21057172 110 - 27905053 AACAA---AAACAAAAACAUUGCCACCGUCUUCAGUGGCUGCUGUUGUCUAUAUACUCUUUAAGUCCAUGAGGGUAUAACAGACUGAUCUUCAUACCUAUU-UUAUAUGGAUGA .....---.......((((..(((((.(....).)))))...))))((((.(((((((((.........)))))))))..))))......((((.((....-......)))))) ( -24.10) >DroSec_CAF1 10544 111 - 1 AACAA---CAACAAAAACAUUGCCACCGUCUUCAGUGGCUGCUGUUGUCUAUAUACUCUUUAAGUCCAUGAGGGUAUAACAGACUGAUCUGCAUACCUAUUUUCAUAUGGAUGA .((((---((.((........(((((.(....).))))))).))))))...............((((((((((((((..((((....)))).))))))....)))..))))).. ( -25.50) >DroSim_CAF1 9751 110 - 1 AACAA---CAACAAAAACAUUGCCACCGUCUUCAGUGGCUGCUGUUGUCUAUAUACUCUUUAAGUCCAUGAGGGUAUAACAGACUGAUCUGCAUACCUAUU-UCAUAUGGAUGA .((((---((.((........(((((.(....).))))))).))))))...............((((((((((((((..((((....)))).))))))...-)))..))))).. ( -25.90) >DroYak_CAF1 10064 108 - 1 AACAACAAAAACAAAAGCAUUGCCACCGUCUUCAGUGGCUGCUGUUGUG-----ACUCUUUGAAUCCAUAGAGGUAUGCUAGACCGAUCUGCAUACCCAUU-UUAUAUGGAAGA .....((((.((((.((((..(((((.(....).))))))))).)))).-----....))))..((((((..(((((((.(((....))))))))))....-...))))))... ( -32.50) >consensus AACAA___AAACAAAAACAUUGCCACCGUCUUCAGUGGCUGCUGUUGUCUAUAUACUCUUUAAGUCCAUGAGGGUAUAACAGACUGAUCUGCAUACCUAUU_UCAUAUGGAUGA ...............((((..(((((.(....).)))))...)))).................(((((((.((((((..((((....)))).)))))).......))))))).. (-19.92 = -20.18 + 0.25)


| Location | 21,057,209 – 21,057,308 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 78.29 |
| Mean single sequence MFE | -29.68 |
| Consensus MFE | -15.07 |
| Energy contribution | -15.99 |
| Covariance contribution | 0.92 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.37 |
| Structure conservation index | 0.51 |
| SVM decision value | 1.43 |
| SVM RNA-class probability | 0.953498 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 21057209 99 + 27905053 UACCCUCAUGGACUUAAAGAGUAUAUAGA-CAACAGCAGCCACUGAAGACGGUGGCAAUGUUUUUGUUU---UUGUUUCCUUGGUUCCGGUAUUUUCGAG---CGA ..(.(((..(((.((((.(((.(((.(((-(((.(((((((((((....)))))))..)))).))))))---.)))))).)))).)))((.....)))))---.). ( -27.70) >DroSec_CAF1 10582 99 + 1 UACCCUCAUGGACUUAAAGAGUAUAUAGA-CAACAGCAGCCACUGAAGACGGUGGCAAUGUUUUUGUUG---UUGUUUCCUUGGUUCCGGUAUUUUUGAG---CCA ........(((.((((((((......(((-(((((((((((((((....)))))))........)))))---))))))((........))..))))))))---))) ( -29.90) >DroSim_CAF1 9788 99 + 1 UACCCUCAUGGACUUAAAGAGUAUAUAGA-CAACAGCAGCCACUGAAGACGGUGGCAAUGUUUUUGUUG---UUGUUUCCUUGGUUCCGGUAUUUUUGAG---CCA ........(((.((((((((......(((-(((((((((((((((....)))))))........)))))---))))))((........))..))))))))---))) ( -29.90) >DroYak_CAF1 10101 97 + 1 UACCUCUAUGGAUUCAAAGAGU-----CA-CAACAGCAGCCACUGAAGACGGUGGCAAUGCUUUUGUUUUUGUUGUUUCCUUGGUUCCGGUAUUUUUGAG---CCA .........(((..(((.(((.-----.(-(((.(((((((((((....)))))))..)))).)))).))).)))..))).((((((..(....)..)))---))) ( -30.50) >DroAna_CAF1 9332 91 + 1 ------------CUCAAAGGGUAUGCAGAAGGACAACAACCACUAAAGGCGGUUGAAAAGUUCUUGUUG---UUGUUUCCUUGGUUCCGGAACACCGGGACUACUA ------------....(((((...((((((((((..(((((.(.....).)))))....)))))).)))---)....)))))((((((((....)))))))).... ( -30.40) >consensus UACCCUCAUGGACUUAAAGAGUAUAUAGA_CAACAGCAGCCACUGAAGACGGUGGCAAUGUUUUUGUUG___UUGUUUCCUUGGUUCCGGUAUUUUUGAG___CCA ........((((.((((.(((.........(((.(((((((((((....)))))))..)))).)))..........))).)))).))))................. (-15.07 = -15.99 + 0.92)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:56:43 2006