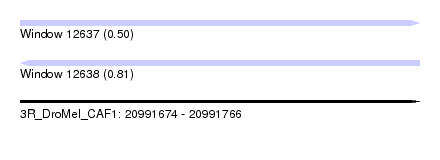
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 20,991,674 – 20,991,766 |
| Length | 92 |
| Max. P | 0.809139 |

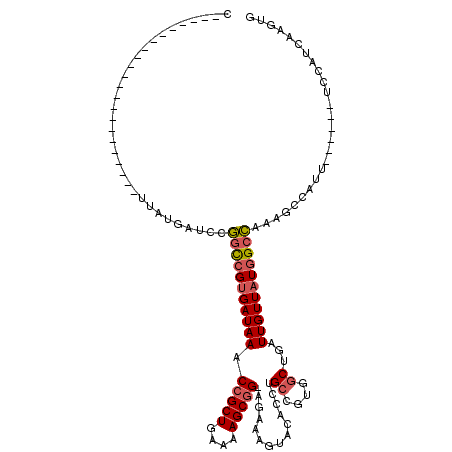
| Location | 20,991,674 – 20,991,766 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.32 |
| Mean single sequence MFE | -26.90 |
| Consensus MFE | -18.08 |
| Energy contribution | -18.22 |
| Covariance contribution | 0.14 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.31 |
| Structure conservation index | 0.67 |
| SVM decision value | -0.07 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 20991674 92 + 27905053 CACUUGAUGGA------AAUGGCUUUGGCCAUAACAAUCAGCCACGGCAGGUGUACUUUCUACCGCUUUUCAGCGGUUUAUCACGGCCGGAUCAUAA----------------------G ...(((((...------.(((((....)))))....)))))...((((..(((........((((((....))))))....))).))))........----------------------. ( -28.10) >DroSec_CAF1 32254 91 + 1 CACUUGAUGGA------AAUGGCUCUGGCCAUAACAAUCAGCCACGGCAGGUGUACUUUCU-CCGCUUUUCAGCGGUUUAUCACGGCCGUAUCAUAA----------------------G ...(((((...------.(((((....)))))....)))))..(((((..(((........-(((((....))))).....))).))))).......----------------------. ( -27.92) >DroSim_CAF1 27664 91 + 1 CACUUGAUGGA------AAUGGCUACGGCCAUAACAAUCAGCCACGGCAGGUGUACUUUCU-CCGCUUUUCAGCGGUUUAUCACGGCCGUAUCAUAA----------------------G ...(((((...------.(((((....)))))....)))))..(((((..(((........-(((((....))))).....))).))))).......----------------------. ( -30.42) >DroEre_CAF1 34237 91 + 1 CACUUGAUGGA------AAUGGCUCCGGCCAUAACAAUCAGCAACGGCAGGUGUACUUACU-CCGCUUUUCAGCGGUUUAUCACGGCCGGAUCAUAC----------------------G ...........------.((((.(((((((..........((....))..(((....((..-(((((....)))))..)).))))))))))))))..----------------------. ( -29.00) >DroYak_CAF1 33822 119 + 1 CACUUGAUGGAAAUGGAAAUGGCUACGGCCAUAACAAUCAGCAACGCCAGGUGUACUUUCU-CCGCUUUUCAGCGGUUUAUCACGACCGGAUUAUACUCACUACUCACUACUCACCACUG ((((((.((....(((..(((((....))))).....)))....)).)))))).......(-(((((....)))((((......)))))))............................. ( -25.10) >DroAna_CAF1 32067 87 + 1 CACUUGAUGGA------AAUGGCUUUUCCCAUAACAAUCAGCCACGGCAGGUGUGU----A-CGGCUUUUCAGCGGUUUAUCGCCGCAGGAUCAUAG----------------------C ..((((.....------..(((((...............)))))((((.((((...----(-(.(((....))).)).)))))))))))).......----------------------. ( -20.86) >consensus CACUUGAUGGA______AAUGGCUACGGCCAUAACAAUCAGCCACGGCAGGUGUACUUUCU_CCGCUUUUCAGCGGUUUAUCACGGCCGGAUCAUAA______________________G ..................((((.(((((((..........((....))..(((.........(((((....))))).....))))))))))))))......................... (-18.08 = -18.22 + 0.14)

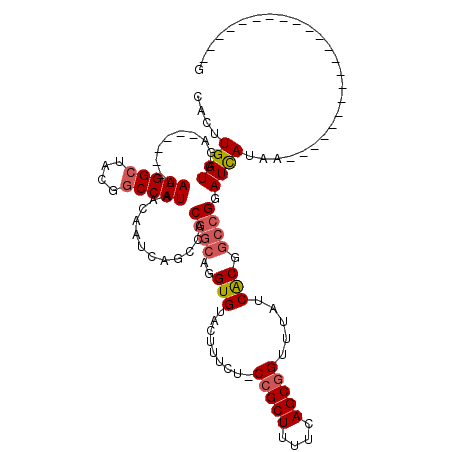
| Location | 20,991,674 – 20,991,766 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.32 |
| Mean single sequence MFE | -31.33 |
| Consensus MFE | -21.61 |
| Energy contribution | -22.03 |
| Covariance contribution | 0.42 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.64 |
| SVM RNA-class probability | 0.809139 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 20991674 92 - 27905053 C----------------------UUAUGAUCCGGCCGUGAUAAACCGCUGAAAAGCGGUAGAAAGUACACCUGCCGUGGCUGAUUGUUAUGGCCAAAGCCAUU------UCCAUCAAGUG .----------------------.........(((((((((((((((((....))))))....(((.(((.....))))))..))))))))))).........------........... ( -30.00) >DroSec_CAF1 32254 91 - 1 C----------------------UUAUGAUACGGCCGUGAUAAACCGCUGAAAAGCGG-AGAAAGUACACCUGCCGUGGCUGAUUGUUAUGGCCAGAGCCAUU------UCCAUCAAGUG .----------------------......((((((.(((((...(((((....)))))-.....)).)))..))))))((((((.(..(((((....))))).------.).))).))). ( -30.40) >DroSim_CAF1 27664 91 - 1 C----------------------UUAUGAUACGGCCGUGAUAAACCGCUGAAAAGCGG-AGAAAGUACACCUGCCGUGGCUGAUUGUUAUGGCCGUAGCCAUU------UCCAUCAAGUG .----------------------..(((.((((((((((((((.(((((....)))))-....(((.(((.....))))))..))))))))))))))..))).------........... ( -34.40) >DroEre_CAF1 34237 91 - 1 C----------------------GUAUGAUCCGGCCGUGAUAAACCGCUGAAAAGCGG-AGUAAGUACACCUGCCGUUGCUGAUUGUUAUGGCCGGAGCCAUU------UCCAUCAAGUG .----------------------..(((.((((((((((((((.(((((....)))))-((((((((....)))..)))))..))))))))))))))..))).------........... ( -36.10) >DroYak_CAF1 33822 119 - 1 CAGUGGUGAGUAGUGAGUAGUGAGUAUAAUCCGGUCGUGAUAAACCGCUGAAAAGCGG-AGAAAGUACACCUGGCGUUGCUGAUUGUUAUGGCCGUAGCCAUUUCCAUUUCCAUCAAGUG ...(((((.(.((((.(.((((.((......((((((((((((.(((((....)))))-....((((.((.....))))))..))))))))))))..)))))).))))).)))))).... ( -34.50) >DroAna_CAF1 32067 87 - 1 G----------------------CUAUGAUCCUGCGGCGAUAAACCGCUGAAAAGCCG-U----ACACACCUGCCGUGGCUGAUUGUUAUGGGAAAAGCCAUU------UCCAUCAAGUG (----------------------(((..((((..(((((....((.(((....))).)-)----.......)))))..)..)))..).(((((((......))------)))))..))). ( -22.60) >consensus C______________________UUAUGAUCCGGCCGUGAUAAACCGCUGAAAAGCGG_AGAAAGUACACCUGCCGUGGCUGAUUGUUAUGGCCAAAGCCAUU______UCCAUCAAGUG ................................(((((((((((.(((((....)))))..............((....))...))))))))))).......................... (-21.61 = -22.03 + 0.42)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:56:07 2006