| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 20,989,367 – 20,989,502 |
| Length | 135 |
| Max. P | 0.997242 |

| Location | 20,989,367 – 20,989,476 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 74.60 |
| Mean single sequence MFE | -39.02 |
| Consensus MFE | -22.42 |
| Energy contribution | -25.22 |
| Covariance contribution | 2.80 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.57 |
| Structure conservation index | 0.57 |
| SVM decision value | 2.82 |
| SVM RNA-class probability | 0.997242 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 20989367 109 - 27905053 AUGUGUA----UGUG--UGUGUACACAAACACA-ACUGUGCGUCGGCUGUAGAGUCGAGGCGAUUCGAGUCCUGGCGUGGUCGUCAGAUGGCUAGGAUGCAGUCUUGAUACAUGAG .(((((.----((((--(....))))).)))))-(((((((.((((((....)))))).)).......((((((((...(((....))).)))))))))))))............. ( -40.60) >DroSec_CAF1 29954 113 - 1 AUAUAUGUGUGUGUG--UGUGUGCACACAUACA-ACUGUGCGUCGGCUGUGGAGUCGAAGCGAUUCGAGUCCUGGCGUGGUCGUCAGAUGGCUAGGAUGCAGUCUUGAUACAUGAG ...((((((((((((--((....))))))))).-....(((((((((((((((.(((((....))))).)))(((((....))))).))))))..))))))........))))).. ( -44.70) >DroSim_CAF1 25382 115 - 1 AUGUGUGUGUGUGUGUUUGUGUGCACACAUACA-ACUGUGCGUCGGCUGUGGAGUCGAGGCGAUUCGAGUCCUGGCGUGGCCGUCAGAUGGCUAGGAUGCAGUCUUGAUACAUGAG ((((((((((((((((......)))))))))).-....((((((.((((.(((.(((((....))))).))))))).(((((((...))))))).))))))......))))))... ( -48.50) >DroWil_CAF1 40408 96 - 1 AUGUAUGCAUGAAUG--UUUGUACAUAUAUACAUACAGAAAGUCGACUGCUU---------------GCUCUUGGCCUGG---GCAGGUGGCUAGAAUGCAGUCUUGAUACAUGAG .((((((.(((.(((--(....)))).))).)))))).......((((((..---------------.((..(.((((..---..)))).)..))...))))))............ ( -26.60) >DroYak_CAF1 31457 93 - 1 AU----------------------ACACAUUCA-ACUGUGCGUCGGCUGUGGAGUCGAGGCGAUUCGAGUCCUGGCCUGGUCGUCAGAUGGCUAGGAUGCAGUCUUGAUACAUGAG ..----------------------...((((((-(((((((.((((((....)))))).)).......(((((((((..(((....)))))))))))))))))..)))...))).. ( -34.70) >consensus AUGUAUG__UGUGUG__UGUGUACACACAUACA_ACUGUGCGUCGGCUGUGGAGUCGAGGCGAUUCGAGUCCUGGCGUGGUCGUCAGAUGGCUAGGAUGCAGUCUUGAUACAUGAG .((((((.(((((........))))).))))))....(((.(((((((((.((((((...))))))..((((((((..............))))))))))))))..))).)))... (-22.42 = -25.22 + 2.80)

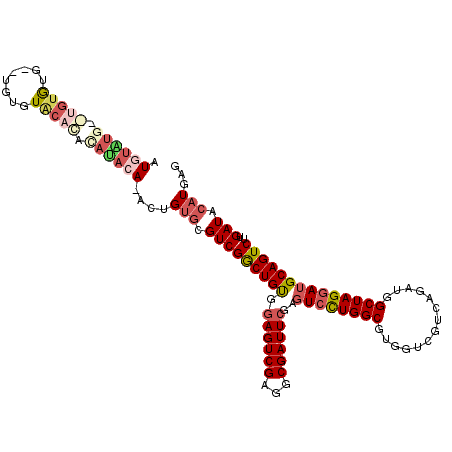

| Location | 20,989,406 – 20,989,502 |
|---|---|
| Length | 96 |
| Sequences | 3 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 90.79 |
| Mean single sequence MFE | -33.30 |
| Consensus MFE | -28.14 |
| Energy contribution | -29.70 |
| Covariance contribution | 1.56 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.566176 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 20989406 96 + 27905053 CGCCAGGACUCGAAUCGCCUCGACUCUACAGCCGACGCACAGUUGUGUUUGUGUACACA--CACA----UACACAUAGGUGCGUGUGUCCGUUCGUGUUCGC .....((((.((((((((..((.((....)).))((((((...(((((.(((((....)--))))----.)))))...)))))))))...))))).)))).. ( -32.90) >DroSec_CAF1 29993 100 + 1 CGCCAGGACUCGAAUCGCUUCGACUCCACAGCCGACGCACAGUUGUAUGUGUGCACACA--CACACACACAUAUAUAGGUGCGUGUGUCCGUUCGUGUUCGC .....((((.((((((((..((.((....)).))((((((...((((((((((......--....))))))))))...)))))))))...))))).)))).. ( -33.00) >DroSim_CAF1 25421 102 + 1 CGCCAGGACUCGAAUCGCCUCGACUCCACAGCCGACGCACAGUUGUAUGUGUGCACACAAACACACACACACACAUAGGUGCGUGUGUCCGUUCGUGUUCGC .....(((.((((......)))).)))(((..((((.....))))..)))(((.((((.(((..((((((.(((....))).))))))..))).)))).))) ( -34.00) >consensus CGCCAGGACUCGAAUCGCCUCGACUCCACAGCCGACGCACAGUUGUAUGUGUGCACACA__CACACACACACACAUAGGUGCGUGUGUCCGUUCGUGUUCGC .....((((.((((((((..((.((....)).))((((((...((((((((((............))))))))))...)))))))))...))))).)))).. (-28.14 = -29.70 + 1.56)

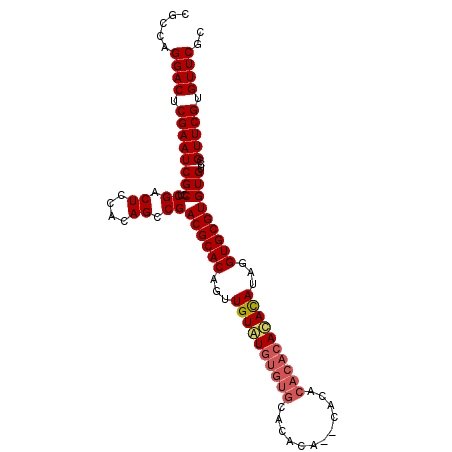

| Location | 20,989,406 – 20,989,502 |
|---|---|
| Length | 96 |
| Sequences | 3 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 90.79 |
| Mean single sequence MFE | -42.37 |
| Consensus MFE | -32.88 |
| Energy contribution | -34.33 |
| Covariance contribution | 1.45 |
| Combinations/Pair | 1.12 |
| Mean z-score | -3.14 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.85 |
| SVM RNA-class probability | 0.866060 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 20989406 96 - 27905053 GCGAACACGAACGGACACACGCACCUAUGUGUA----UGUG--UGUGUACACAAACACAACUGUGCGUCGGCUGUAGAGUCGAGGCGAUUCGAGUCCUGGCG ((.(((.((((((((((((..(((....)))..----))))--)((((......))))..))))((.((((((....)))))).))...))).))..).)). ( -33.40) >DroSec_CAF1 29993 100 - 1 GCGAACACGAACGGACACACGCACCUAUAUAUGUGUGUGUG--UGUGUGCACACAUACAACUGUGCGUCGGCUGUGGAGUCGAAGCGAUUCGAGUCCUGGCG .......((((((.((((((((((........)))))))))--).)))(((((........))))).)))((((.(((.(((((....))))).))))))). ( -42.20) >DroSim_CAF1 25421 102 - 1 GCGAACACGAACGGACACACGCACCUAUGUGUGUGUGUGUGUUUGUGUGCACACAUACAACUGUGCGUCGGCUGUGGAGUCGAGGCGAUUCGAGUCCUGGCG .((((((((((((.((((((((((....)))))))))).)))))))))(((((........))))).)))((((.(((.(((((....))))).))))))). ( -51.50) >consensus GCGAACACGAACGGACACACGCACCUAUGUGUGUGUGUGUG__UGUGUGCACACAUACAACUGUGCGUCGGCUGUGGAGUCGAGGCGAUUCGAGUCCUGGCG ((........((((...(((((((...((((((((((((........))))))))))))...)))))).).))))(((.(((((....))))).)))..)). (-32.88 = -34.33 + 1.45)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:56:02 2006