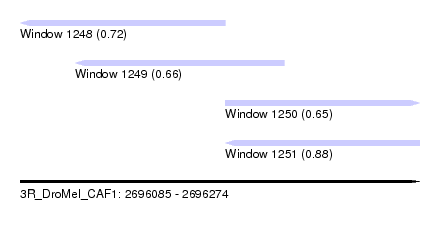
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 2,696,085 – 2,696,274 |
| Length | 189 |
| Max. P | 0.879920 |

| Location | 2,696,085 – 2,696,182 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 88.21 |
| Mean single sequence MFE | -34.72 |
| Consensus MFE | -28.81 |
| Energy contribution | -29.45 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.41 |
| SVM RNA-class probability | 0.724171 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 2696085 97 - 27905053 --GCCAUCCACUGAAUCCGAGGAUGCAUAUGCAUCCUCAUAUGUUUUUUCGGCGGCCUCUUUGACACGAAAAUCCGCGAUGGAUGCCGGUCCUCUUCGG- --..((((((.((.....((((((((....))))))))....(..((((((.(((.....)))...))))))..).)).))))))((((......))))- ( -31.60) >DroSec_CAF1 6050 97 - 1 --GCCAUCCGCUGAAUCCGAGGAUGCAUAUGCAUCCUCACAUGUUGUUUCGGCGGCCUCUUUGACACGAAAAUCCGCGGUGGAUGCCGGUCCUCUUCGG- --..(((((((((.....((((((((....))))))))........(((((.(((.....)))...))))).....)))))))))((((......))))- ( -34.90) >DroSim_CAF1 6063 96 - 1 --GCCAUCCGCUGAAUCCGAGGAUGCAUAUGCAUCCUCACAUGUUGUUUCGGCGGCCUCUUUGACACGAAAAUCCGCG-UGGAUGCCGGUCCUCUUCGG- --.((..((((((((...((((((((....))))))))((.....)))))))))).......(((.((...((((...-.))))..)))))......))- ( -34.00) >DroEre_CAF1 7008 96 - 1 --GCCAUCCGCUGAAUCCGAGGAUGCAUAUGCAUCCUCACAUGUU-UUUCGGCGGCCUCUUUGACAAGAAUAUCCGCGGUGGAUGCCGGUCCUCUUCGG- --..((((((((......((((((((....)))))))).......-.....((((..((((....))))....))))))))))))((((......))))- ( -36.60) >DroYak_CAF1 7209 96 - 1 --GCCAUCCGCUGAAUCCGAGGAUGCAUAUGCAUCCUCACAUGUU-UUUCGUCGGCCUCUUUGACACGAAAAUCUGCGGUGGAUGCUGGCCCUCUUCGG- --(((((((((((.....((((((((....)))))))).((.(((-(((.(((((.....)))))..)))))).)))))))))...)))).........- ( -35.00) >DroAna_CAF1 6944 93 - 1 GUGCCAUCCGCGGAUUCUGAGGAUGU------CUGCUCAUGUGUU-UUUCGGCGGUCUCUUUGACUCGAAAAUCCGCGGUGGAUGCCGCUCGUCGGCGGU ...(((.(((((((...((((.....------...)))).....(-(((((..((((.....))))))))))))))))))))..((((((....)))))) ( -36.20) >consensus __GCCAUCCGCUGAAUCCGAGGAUGCAUAUGCAUCCUCACAUGUU_UUUCGGCGGCCUCUUUGACACGAAAAUCCGCGGUGGAUGCCGGUCCUCUUCGG_ ....((((((((......((((((((....)))))))).............((((..((........))....))))))))))))((((......)))). (-28.81 = -29.45 + 0.64)

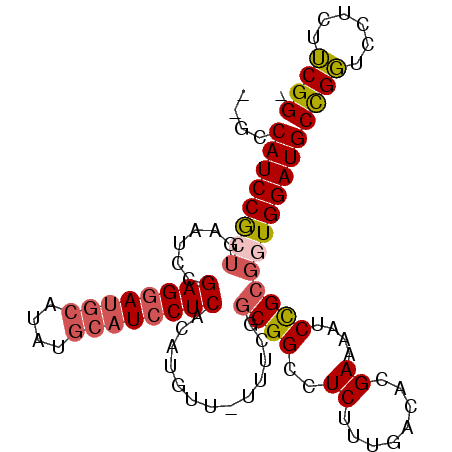
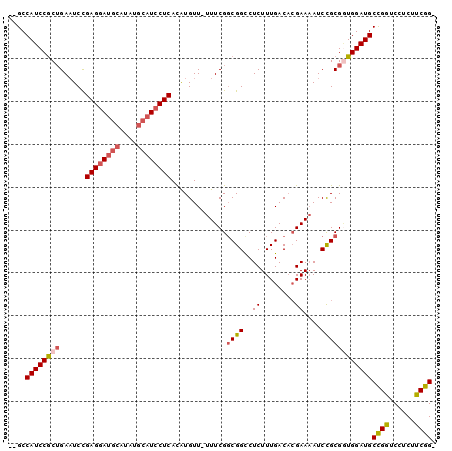
| Location | 2,696,111 – 2,696,210 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 82.45 |
| Mean single sequence MFE | -32.45 |
| Consensus MFE | -22.64 |
| Energy contribution | -23.17 |
| Covariance contribution | 0.53 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.655299 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 2696111 99 - 27905053 GCUCUCAAGAUGUGCUACGUCGAGCGGUGCCAUCCACUGA-----AUCCGAGGAUGCAUAUGCAUCCUCAUAUGUUU--UUUCGGCGGCCUCUUUGACACGAAAAU ....(((((..(.((..((((((((((((.....))))).-----....((((((((....))))))))........--.))))))))))..)))))......... ( -31.80) >DroPse_CAF1 36088 103 - 1 CCACUCAAGAUGUGCUACGUCGAGCGGUGCAACGUUUUGUGUGUCGUCCGUGGAUGCCU---CCUCCUCACAUGGCGGUUUUUGGCGGUCUCUUUGACACAAAAUU .....((((((((((..((.....))..)).))))))))(((((((.((((.((.(((.---((.........)).)))..)).))))......)))))))..... ( -31.60) >DroSim_CAF1 6088 99 - 1 GUUCUCAAGAUGUGCUACGUCGAGCGGUGCCAUCCGCUGA-----AUCCGAGGAUGCAUAUGCAUCCUCACAUGUUG--UUUCGGCGGCCUCUUUGACACGAAAAU ((..(((((..(.((..((((((((((......))))...-----....((((((((....))))))))........--..)))))))))..))))).))...... ( -34.30) >DroEre_CAF1 7034 98 - 1 CCUCUCAAGAUGUGCUACGUCGAGCGGUGCCAUCCGCUGA-----AUCCGAGGAUGCAUAUGCAUCCUCACAUGUU---UUUCGGCGGCCUCUUUGACAAGAAUAU ....(((((..(.((..((((((((((......))))...-----....((((((((....)))))))).......---..)))))))))..)))))......... ( -33.90) >DroYak_CAF1 7235 98 - 1 CCUCUCAAGAUGUGCUACGUCGAGCGGUGCCAUCCGCUGA-----AUCCGAGGAUGCAUAUGCAUCCUCACAUGUU---UUUCGUCGGCCUCUUUGACACGAAAAU ....(((((..(.((((((((.(((((......)))))))-----....((((((((....)))))))).......---...))).))))..)))))......... ( -31.50) >DroPer_CAF1 36103 103 - 1 CCACUCAAGAUGUGCUACGUCGAGCGGUGCAACGUUUUGUGUGUCGUCCGUGGAUGCCU---CCUCCUCACAUGGCGGUUUUUGGCGGUCUCUUUGACACAAAAUU .....((((((((((..((.....))..)).))))))))(((((((.((((.((.(((.---((.........)).)))..)).))))......)))))))..... ( -31.60) >consensus CCUCUCAAGAUGUGCUACGUCGAGCGGUGCCAUCCGCUGA_____AUCCGAGGAUGCAUAUGCAUCCUCACAUGUUG__UUUCGGCGGCCUCUUUGACACGAAAAU ....(((((..(.((..((((((((((((.....)))))..........((((((((....))))))))...........))))))))))..)))))......... (-22.64 = -23.17 + 0.53)
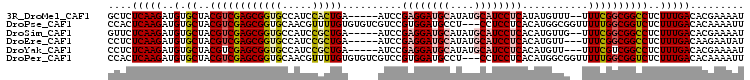
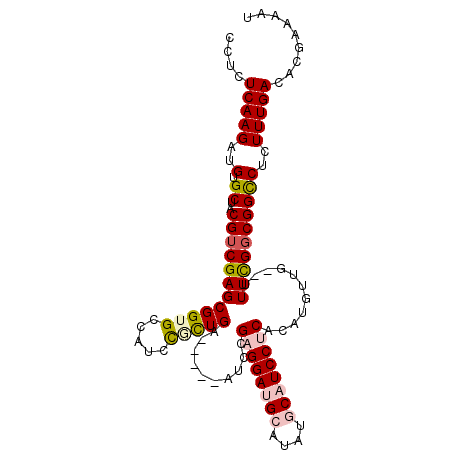
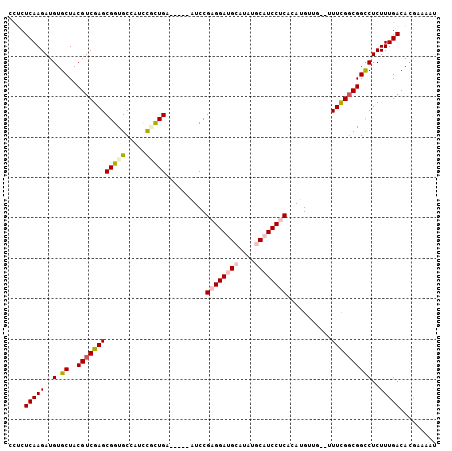
| Location | 2,696,182 – 2,696,274 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 92 |
| Reading direction | forward |
| Mean pairwise identity | 93.70 |
| Mean single sequence MFE | -23.18 |
| Consensus MFE | -18.82 |
| Energy contribution | -19.62 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.645456 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 2696182 92 + 27905053 ACCGCUCGACGUAGCACAUCUUGAGAGCUAUAGAACCACUACCACCGCCUCCGCAUCGGCGGUAUUUAGCUUUAGCUAUAUCACGCAUUAUU ...((..((..((((.(.....)((((((((((.....)))..((((((........))))))...))))))).))))..))..))...... ( -25.10) >DroSec_CAF1 6147 92 + 1 ACCGCUCGACGUAGCACAUCUUGAGAACUAGAGAGCCACUACCACCGCCUCCGCAUCGGCGGUAUUUAGCUUUAGCUAUAUCACGCAUUAUA ...((..((..((((...((....)).....(((((.......((((((........)))))).....))))).))))..))..))...... ( -23.20) >DroSim_CAF1 6159 92 + 1 ACCGCUCGACGUAGCACAUCUUGAGAACUAGAGAACCACUACCACCGCCUCCGCAUCGGCGAUAUUUAGCUUUAGCUAUAUCACGCUUUAUA ...((..((..((((...((((........))))...........((((........)))).............))))..))..))...... ( -17.50) >DroEre_CAF1 7104 92 + 1 ACCGCUCGACGUAGCACAUCUUGAGAGGUAGAGAACCACCAGCACCGCCUCCGCAUCGGCGGAAUUUAGCUUUAGCUAUAUCACGCAUUAUA (((.(((((.((.....)).))))).)))............((.(((((........)))))....((((....))))......))...... ( -23.10) >DroYak_CAF1 7305 92 + 1 ACCGCUCGACGUAGCACAUCUUGAGAGGUAGAGUGCCACUACUACCGCCUCCGCAUCGGCGGUACUUAGCUUUAGCUAUAUCACGCAUUAUA (((.(((((.((.....)).))))).)))..(((((......(((((((........)))))))..((((....))))......)))))... ( -27.00) >consensus ACCGCUCGACGUAGCACAUCUUGAGAGCUAGAGAACCACUACCACCGCCUCCGCAUCGGCGGUAUUUAGCUUUAGCUAUAUCACGCAUUAUA ...((..((..((((...((((........)))).........((((((........))))))...........))))..))..))...... (-18.82 = -19.62 + 0.80)

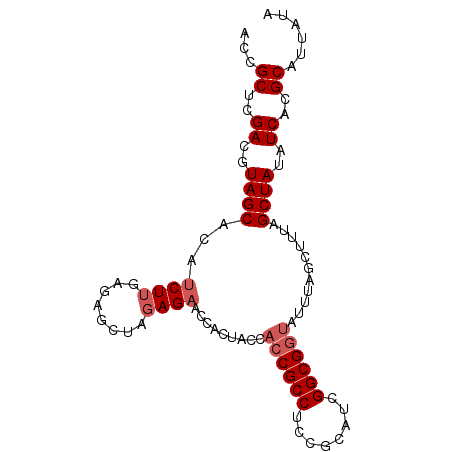
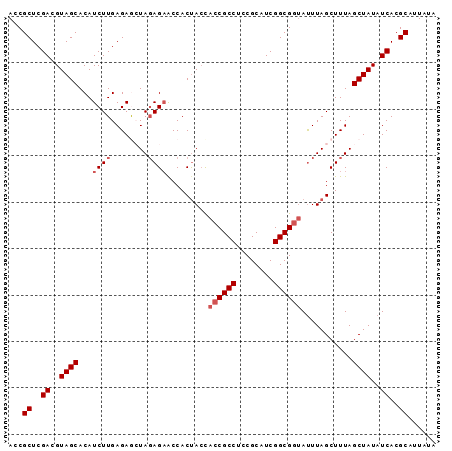
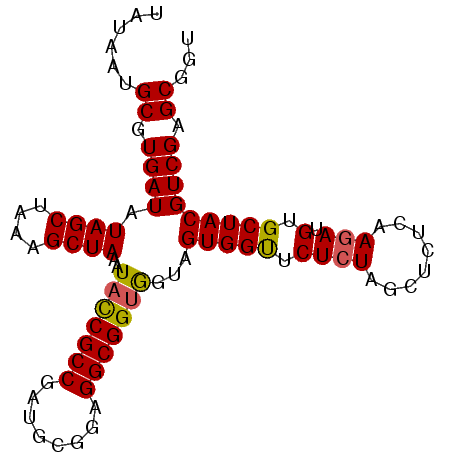
| Location | 2,696,182 – 2,696,274 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 92 |
| Reading direction | reverse |
| Mean pairwise identity | 93.70 |
| Mean single sequence MFE | -29.22 |
| Consensus MFE | -26.16 |
| Energy contribution | -26.00 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.40 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.91 |
| SVM RNA-class probability | 0.879920 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 2696182 92 - 27905053 AAUAAUGCGUGAUAUAGCUAAAGCUAAAUACCGCCGAUGCGGAGGCGGUGGUAGUGGUUCUAUAGCUCUCAAGAUGUGCUACGUCGAGCGGU .....(((.(((...(((((.(((((..(((((((........)))))))....)))))...))))).))).((((.....))))..))).. ( -28.00) >DroSec_CAF1 6147 92 - 1 UAUAAUGCGUGAUAUAGCUAAAGCUAAAUACCGCCGAUGCGGAGGCGGUGGUAGUGGCUCUCUAGUUCUCAAGAUGUGCUACGUCGAGCGGU .....(((.((((.((((....))))..(((((((........)))))))...(((((.((((........))).).))))))))).))).. ( -29.90) >DroSim_CAF1 6159 92 - 1 UAUAAAGCGUGAUAUAGCUAAAGCUAAAUAUCGCCGAUGCGGAGGCGGUGGUAGUGGUUCUCUAGUUCUCAAGAUGUGCUACGUCGAGCGGU ......((.((((.((((....))))..(((((((........)))))))...(((((.((((........))).).))))))))).))... ( -24.90) >DroEre_CAF1 7104 92 - 1 UAUAAUGCGUGAUAUAGCUAAAGCUAAAUUCCGCCGAUGCGGAGGCGGUGCUGGUGGUUCUCUACCUCUCAAGAUGUGCUACGUCGAGCGGU .....(((.(((..((((...(((......(((((........))))).)))(((((....)))))...........))))..))).))).. ( -28.80) >DroYak_CAF1 7305 92 - 1 UAUAAUGCGUGAUAUAGCUAAAGCUAAGUACCGCCGAUGCGGAGGCGGUAGUAGUGGCACUCUACCUCUCAAGAUGUGCUACGUCGAGCGGU .....(((.((((.((((....))))(.(((((((........))))))).).((((((((((........))).))))))))))).))).. ( -34.50) >consensus UAUAAUGCGUGAUAUAGCUAAAGCUAAAUACCGCCGAUGCGGAGGCGGUGGUAGUGGUUCUCUAGCUCUCAAGAUGUGCUACGUCGAGCGGU ......((.((((.((((....))))..(((((((........)))))))...(((((.((((........))).).))))))))).))... (-26.16 = -26.00 + -0.16)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:52:04 2006