
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 20,975,658 – 20,975,753 |
| Length | 95 |
| Max. P | 0.998784 |

| Location | 20,975,658 – 20,975,753 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 71.65 |
| Mean single sequence MFE | -45.60 |
| Consensus MFE | -24.69 |
| Energy contribution | -25.93 |
| Covariance contribution | 1.24 |
| Combinations/Pair | 1.17 |
| Mean z-score | -3.18 |
| Structure conservation index | 0.54 |
| SVM decision value | 3.22 |
| SVM RNA-class probability | 0.998784 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
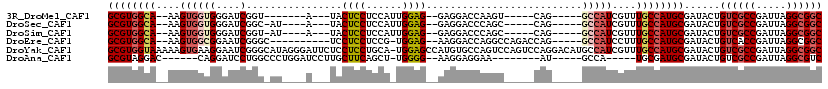
>3R_DroMel_CAF1 20975658 95 + 27905053 GCGUGGCA--AAGUGGUGGGAUCGGU-------A---UACUCCUCCAUUGGAG--GAGGACCAAGU-----CAG-----GCCAUCGUUUGCCAUGCGAUACUGUCGCCGAUUAGGCGGC ((((((((--(((((((..(((.(((-------.---..(((((((...))))--))).)))..))-----)..-----)))))..))))))))))......((((((.....)))))) ( -50.30) >DroSec_CAF1 15700 97 + 1 GCGUGGCA--AAGUGGUGGGAUCGGC-AU----A---UACUCCUCCAUUGGAG--GAGGACCCAGC-----CAG-----GCCAUCGUUUGCCAUGCGAUACUGUCGCCGAUUAGGCGGC ((((((((--((.((((((..(.(((-..----.---..(((((((...))))--)))......))-----)).-----.))))))))))))))))......((((((.....)))))) ( -50.00) >DroSim_CAF1 15763 97 + 1 GCGUGGCA--AAGUGGUGGGAUCGGU-AU----A---UACUCCUCCAUUGGAG--GAGGACCCAGC-----CAG-----GCCAUCGUUUGCCAUGCGAUACUGUCGCCGAUUAGGCGGC ((((((((--(((((((.((...(((-..----.---..(((((((...))))--))).)))...)-----)..-----)))))..))))))))))......((((((.....)))))) ( -49.50) >DroEre_CAF1 17900 99 + 1 GCGUGGCA--AAGUGGCGGAAUCGGGC----------UCCUCCUCCG-UGGAG--AAGGACCAGGCCAGACCAG-----GCCAUCCUUUGCCAUGCGAUACUGUCACCGAUUAGGCGGC ((((((((--(((.((........((.----------((((.(((..-..)))--.)))))).((((......)-----))).)))))))))))))....(((((........))))). ( -43.60) >DroYak_CAF1 16643 118 + 1 GCGUGGUAAAAAGUGAAGGAAUCGGGCAUAGGGAUUCUCCUCCUGCA-UGGAGCCAUGUGCCAGUCCAGUCCAGGACAUGCCAUCGUUUGCCAUGCGAUACUGUCGCCGAUUAGGCGGC ((((((((((..(((.........((((((.((...((((.......-.)))))).)))))).((((......))))....)))..))))))))))......((((((.....)))))) ( -49.50) >DroAna_CAF1 15642 92 + 1 GCGUAGGAC------CAGGAUCCUGGCCCUGGAUCCUUGCUUCAGCU-UGGGG--AAGGAGGAA--------AU-----GCCA-----UGCGAUGCGAUACUGUCGCCGAUUAGGCGUC ...((((.(------(((....)))).))))..(((((.((((....-....)--)))))))).--------((-----(((.-----..((..(((((...)))))))....))))). ( -30.70) >consensus GCGUGGCA__AAGUGGUGGAAUCGGG_AU____A___UACUCCUCCA_UGGAG__AAGGACCAAGC_____CAG_____GCCAUCGUUUGCCAUGCGAUACUGUCGCCGAUUAGGCGGC ((((((((....((((((....)................((((......))))..........................)))))....))))))))......((((((.....)))))) (-24.69 = -25.93 + 1.24)


| Location | 20,975,658 – 20,975,753 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 71.65 |
| Mean single sequence MFE | -36.40 |
| Consensus MFE | -13.88 |
| Energy contribution | -14.97 |
| Covariance contribution | 1.08 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.38 |
| SVM decision value | -0.05 |
| SVM RNA-class probability | 0.505793 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 20975658 95 - 27905053 GCCGCCUAAUCGGCGACAGUAUCGCAUGGCAAACGAUGGC-----CUG-----ACUUGGUCCUC--CUCCAAUGGAGGAGUA---U-------ACCGAUCCCACCACUU--UGCCACGC ..((((.....))))........((.(((((((.(.(((.-----..(-----(.(((((.(((--((((...)))))))..---.-------)))))))...))))))--))))).)) ( -36.10) >DroSec_CAF1 15700 97 - 1 GCCGCCUAAUCGGCGACAGUAUCGCAUGGCAAACGAUGGC-----CUG-----GCUGGGUCCUC--CUCCAAUGGAGGAGUA---U----AU-GCCGAUCCCACCACUU--UGCCACGC ..((((.....))))........((.(((((((.(.(((.-----.((-----((...((.(((--((((...))))))).)---)----..-))))...))).)..))--))))).)) ( -37.00) >DroSim_CAF1 15763 97 - 1 GCCGCCUAAUCGGCGACAGUAUCGCAUGGCAAACGAUGGC-----CUG-----GCUGGGUCCUC--CUCCAAUGGAGGAGUA---U----AU-ACCGAUCCCACCACUU--UGCCACGC ..((((.....))))........((.(((((((.(.(((.-----..(-----(.(.(((.(((--((((...)))))))..---.----..-))).).))..))))))--))))).)) ( -35.30) >DroEre_CAF1 17900 99 - 1 GCCGCCUAAUCGGUGACAGUAUCGCAUGGCAAAGGAUGGC-----CUGGUCUGGCCUGGUCCUU--CUCCA-CGGAGGAGGA----------GCCCGAUUCCGCCACUU--UGCCACGC ..((((.....))))........((.((((((((..((((-----..((((.(((....(((((--(((..-..))))))))----------))).))))..)))))))--))))).)) ( -44.30) >DroYak_CAF1 16643 118 - 1 GCCGCCUAAUCGGCGACAGUAUCGCAUGGCAAACGAUGGCAUGUCCUGGACUGGACUGGCACAUGGCUCCA-UGCAGGAGGAGAAUCCCUAUGCCCGAUUCCUUCACUUUUUACCACGC ..((((.....))))...(((..((((((....(.(((((..((((......))))..)).))).)..)))-)))..((((.(((((.........)))))))))......)))..... ( -35.40) >DroAna_CAF1 15642 92 - 1 GACGCCUAAUCGGCGACAGUAUCGCAUCGCA-----UGGC-----AU--------UUCCUCCUU--CCCCA-AGCUGAAGCAAGGAUCCAGGGCCAGGAUCCUG------GUCCUACGC ...(((......((((.....))))......-----.)))-----..--------.((((.(((--(....-....))))..))))...((((((((....)))------))))).... ( -30.32) >consensus GCCGCCUAAUCGGCGACAGUAUCGCAUGGCAAACGAUGGC_____CUG_____GCUUGGUCCUC__CUCCA_UGGAGGAGUA___U____AU_CCCGAUCCCACCACUU__UGCCACGC ..((((.....))))........((.(((((.....(((...........................((((......))))................(....).))).....))))).)) (-13.88 = -14.97 + 1.08)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:55:42 2006