
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 20,947,733 – 20,947,884 |
| Length | 151 |
| Max. P | 0.856488 |

| Location | 20,947,733 – 20,947,845 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.24 |
| Mean single sequence MFE | -27.53 |
| Consensus MFE | -20.37 |
| Energy contribution | -19.85 |
| Covariance contribution | -0.52 |
| Combinations/Pair | 1.35 |
| Mean z-score | -1.33 |
| Structure conservation index | 0.74 |
| SVM decision value | -0.00 |
| SVM RNA-class probability | 0.531768 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
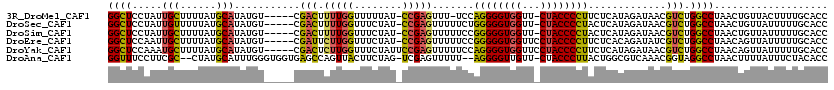
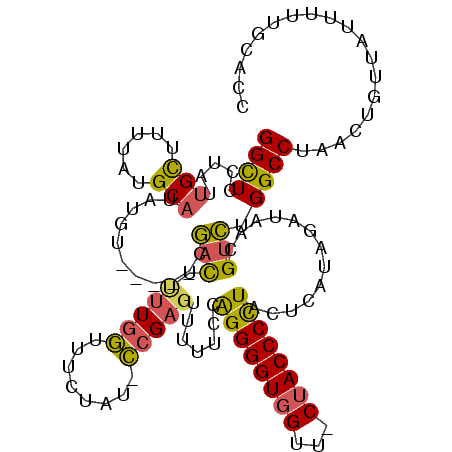
>3R_DroMel_CAF1 20947733 112 - 27905053 GGCUCCUAUUGCUUUUAUGCAUAUGU-----CGACUUUUGGUUUUUAU-CCGAGUUU-UCCAGGGGUGGUU-CUACCCCUUCUCAUAGAUAACGUCUGGCCUAACUGUUACUUUUGCACC (((.......)))....((((...((-----.(((....((((.((((-(.(((...-...(((((((...-.))))))).)))...))))).....)))).....)))))...)))).. ( -24.30) >DroSec_CAF1 11175 113 - 1 GGCUCCUAUUGUUUUUAUGCAUAUGU-----CGACUUUUGGUUUCUAU-CCGAGUUUUUCUGGGGGUGGUU-CUACCCCUACUCAUAGAUAACGUCUGGCCUAACUGUUAUUUUUGCACC ((((.....((((((((((.......-----.....(((((.......-)))))......((((((((...-.))))))))..)))))).))))...))))................... ( -24.20) >DroSim_CAF1 11035 113 - 1 GGCUCCUAUUGCUUUUAUGCAUAUGU-----CGACUUUUGGUUUCUAU-CCGAGUUUUUCCGGGGGUGGUU-CUACCCCUACUCAUAGAUAACGUCUGGCCUAACUGUUAUUUUUGCACC ((((.....(((......))).....-----.(((.....((((((((-..((((......(((((((...-.)))))))))))))))).)))))).))))................... ( -25.50) >DroEre_CAF1 11087 114 - 1 GGCUCCAAUUGCUUUUAUGCAUAUGU-----CGAUUCUUGGUUUCUAU-CCGAGUUUUUCCGGGGGUGGUUCCUACCCCUUCUCACAGAUAUCGUCUGGCCUAACAGUUAUUUUUGCACC .((...(((.(((.(((.((....(.-----.((..(((((.......-)))))..))..)((((((((...)))))))).....(((((...))))))).))).))).)))...))... ( -28.40) >DroYak_CAF1 11175 115 - 1 GGCUCCAAAUGCUUUUAUGCAUAUGU-----CGACUCUUGGUUUCUAUUCCGAGUUUUUCCAGGGGUGGUUCCUACCCCUUCUCAUAGAUAACGUCUGGCCUAACAGUUAUUUUUGCACC ((((.((.((((......)))).)).-----.(((.(((((........))))).......((((((((...)))))))).............))).))))................... ( -28.90) >DroAna_CAF1 11843 114 - 1 GGUUUCCUUCGC--CUAUGCAUUUGGGUGGUGAGCCAGUUACUUCUAG-UCGAGUUUUU--AGGGGUUGUU-CUACCCUUACUGGCGUCAAACGGUAGGCCUAACUUUUAUUUCUACACC (((((((.((((--(((......)))))))((((((((..((((....-..))))...(--((((((....-..)))))))))))).)))...)).)))))................... ( -33.90) >consensus GGCUCCUAUUGCUUUUAUGCAUAUGU_____CGACUUUUGGUUUCUAU_CCGAGUUUUUCCAGGGGUGGUU_CUACCCCUACUCAUAGAUAACGUCUGGCCUAACUGUUAUUUUUGCACC ((((.....(((......)))...........(((.(((((........))))).......((((((((...)))))))).............))).))))................... (-20.37 = -19.85 + -0.52)

| Location | 20,947,773 – 20,947,884 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.09 |
| Mean single sequence MFE | -32.47 |
| Consensus MFE | -23.54 |
| Energy contribution | -23.10 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.44 |
| Mean z-score | -2.03 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.81 |
| SVM RNA-class probability | 0.856488 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
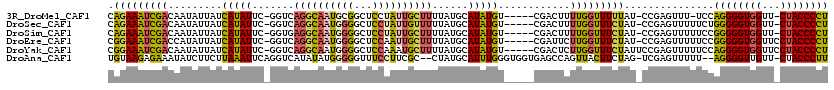
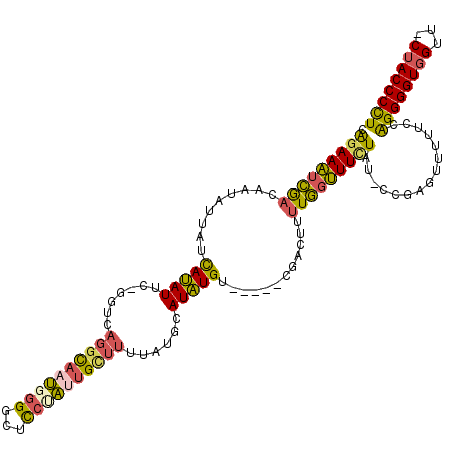
>3R_DroMel_CAF1 20947773 111 - 27905053 CAGAAAUCGACAAUAUUAUCAUAUUC-GGUCAGGCAAUGCGGCUCCUAUUGCUUUUAUGCAUAUGU-----CGACUUUUGGUUUUUAU-CCGAGUUU-UCCAGGGGUGGUU-CUACCCCU (((((.((((((....(((((((...-....((((((((.(....))))))))).)))).))))))-----))).)))))........-........-...(((((((...-.))))))) ( -28.20) >DroSec_CAF1 11215 112 - 1 CAGAAAUCGACAAUAUUAUCAUAUUC-GGUCAGGCAAUGGGGCUCCUAUUGUUUUUAUGCAUAUGU-----CGACUUUUGGUUUCUAU-CCGAGUUUUUCUGGGGGUGGUU-CUACCCCU ((((((((((((....(((((((...-....((((((((((...)))))))))).)))).))))))-----)))..(((((.......-)))))..))))))((((((...-.)))))). ( -32.80) >DroSim_CAF1 11075 112 - 1 CAGAAAUCGACAAUAUUAUCAUAUUC-GGUGAGGCAAUGGGGCUCCUAUUGCUUUUAUGCAUAUGU-----CGACUUUUGGUUUCUAU-CCGAGUUUUUCCGGGGGUGGUU-CUACCCCU (((((.((((((...(((((......-)))))(((((((((...)))))))))..........)))-----))).)))))........-............(((((((...-.))))))) ( -34.60) >DroEre_CAF1 11127 113 - 1 CGGAAAUCGACCAUAUUAUCAUAUUC-GGUCAGGCAAUGGGGCUCCAAUUGCUUUUAUGCAUAUGU-----CGAUUCUUGGUUUCUAU-CCGAGUUUUUCCGGGGGUGGUUCCUACCCCU (((((((((((.((((...((((...-....(((((((((....)).))))))).)))).))))))-----)))..(((((.......-)))))..))))))(((((((...))))))). ( -36.10) >DroYak_CAF1 11215 114 - 1 CGGAAAUCGACAAUAUUAUCAUAUUC-GGUCAGGCAAUGGGGCUCCAAAUGCUUUUAUGCAUAUGU-----CGACUCUUGGUUUCUAUUCCGAGUUUUUCCAGGGGUGGUUCCUACCCCU .(((((((((((..............-((...(((......)))))..((((......)))).)))-----)))..(((((........)))))..)))))((((((((...)))))))) ( -35.90) >DroAna_CAF1 11883 114 - 1 UGUAAGAGAAAUAUCUUCUUAAAUUCAGGUCAUAUAUGGGGGUUUCCUUCGC--CUAUGCAUUUGGGUGGUGAGCCAGUUACUUCUAG-UCGAGUUUUU--AGGGGUUGUU-CUACCCUU ..(((((((....)).)))))................((((((((...((((--(((......))))))).)))))((..((((((((-........))--))))))....-)).))).. ( -27.20) >consensus CAGAAAUCGACAAUAUUAUCAUAUUC_GGUCAGGCAAUGGGGCUCCUAUUGCUUUUAUGCAUAUGU_____CGACUUUUGGUUUCUAU_CCGAGUUUUUCCAGGGGUGGUU_CUACCCCU .(((((((((.........(((((.......((((((((((...))))))))))......)))))............)))))))))...............((((((((...)))))))) (-23.54 = -23.10 + -0.44)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:55:30 2006