
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 20,942,815 – 20,942,933 |
| Length | 118 |
| Max. P | 0.996022 |

| Location | 20,942,815 – 20,942,933 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.18 |
| Mean single sequence MFE | -27.22 |
| Consensus MFE | -21.10 |
| Energy contribution | -22.90 |
| Covariance contribution | 1.80 |
| Combinations/Pair | 1.11 |
| Mean z-score | -3.44 |
| Structure conservation index | 0.78 |
| SVM decision value | 2.64 |
| SVM RNA-class probability | 0.996022 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
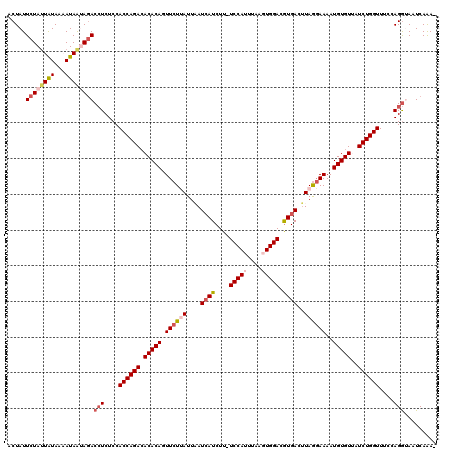
>3R_DroMel_CAF1 20942815 118 + 27905053 ACUAUUCUAUUAUAAAAUAAUAGACCUCUCCACCAGACACACAGUUCUUAUUAAUCAUCUU-UCCAUUUAAGUGGACAUGACUUAAGAAAAUGUGUUAUCUGGUUUCCAGGUAAUCAAA- ......(((((((...)))))))((((....((((((.(((((.((((((....((((...-(((((....))))).))))..))))))..)))))..))))))....)))).......- ( -30.40) >DroSec_CAF1 5967 118 + 1 ACUAUUCUAUUAUAAAAUAAUAUACCUCUCCACCAGACACACAGUUCUUAUUAAUAAUCUU-UCCAUUUAAGUGGACGUGACUUAGGAAAAUGUGUUAUCUGGUUUCCAGGCAAUCAAA- ........................(((....((((((.(((((.((((((.......((..-(((((....)))))...))..))))))..)))))..))))))....)))........- ( -22.10) >DroSim_CAF1 5949 118 + 1 ACUAUUCUAUUAUAAAAUAAUAGACCUCUCCACCAGACACACAGUUCUUAUUAAUCAUCUU-UCCAUUUAAGUGGACGUGACUUAGGAAAAUGUGUUAUCUGGUUUCCAGGCAAUCAAA- .....((((((((...))))))))(((....((((((.(((((.((((((....((((...-(((((....))))).))))..))))))..)))))..))))))....)))........- ( -29.90) >DroEre_CAF1 5863 120 + 1 ACUAUUCUAAUGUAACAUAAAAGACAUCUCCACCAGAAACACAGUUCUAAUUUAUCAUCUUUUCCAUUUAACUGGACGUGACAUAGGAAAAUGUGUAAUCUGGUUUCGAGAUAAUCAGAA .........................(((((.((((((.(((((.(((((.....((((....((((......)))).))))..)))))...)))))..))))))...)))))........ ( -25.00) >DroYak_CAF1 5954 119 + 1 ACUAUUCUAUUAUAACAUAGGAGACCUCUCCACCAGACACACAGUUCUAAUUUAUCAUCUUUUCCAUUAAACUGGACGUGACUUAGUAAAAUGUGUAAUCUGGUUUCGAGGUAAUCAGA- ....((((((......)))))).(((((...((((((.(((((...((((....((((....((((......)))).)))).)))).....)))))..))))))...))))).......- ( -28.70) >consensus ACUAUUCUAUUAUAAAAUAAUAGACCUCUCCACCAGACACACAGUUCUUAUUAAUCAUCUU_UCCAUUUAAGUGGACGUGACUUAGGAAAAUGUGUUAUCUGGUUUCCAGGUAAUCAAA_ .....((((((((...))))))))(((....((((((.(((((.((((((....((((....(((((....))))).))))..))))))..)))))..))))))....)))......... (-21.10 = -22.90 + 1.80)


| Location | 20,942,815 – 20,942,933 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.18 |
| Mean single sequence MFE | -31.66 |
| Consensus MFE | -24.82 |
| Energy contribution | -25.06 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.98 |
| Structure conservation index | 0.78 |
| SVM decision value | 2.41 |
| SVM RNA-class probability | 0.993594 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
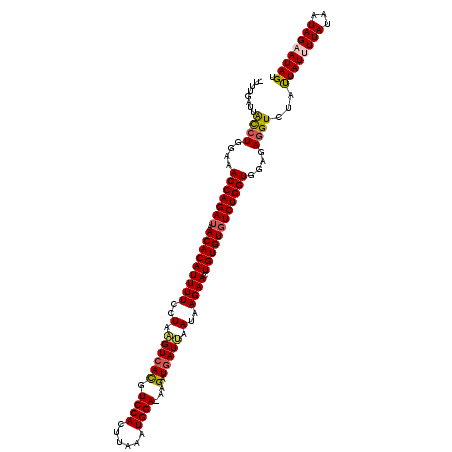
>3R_DroMel_CAF1 20942815 118 - 27905053 -UUUGAUUACCUGGAAACCAGAUAACACAUUUUCUUAAGUCAUGUCCACUUAAAUGGA-AAGAUGAUUAAUAAGAACUGUGUGUCUGGUGGAGAGGUCUAUUAUUUUAUAAUAGAAUAGU -........(((....((((((((.((((..((((((((((((.((((......))))-...))))))..)))))).))))))))))))....)))((((((((...))))))))..... ( -34.90) >DroSec_CAF1 5967 118 - 1 -UUUGAUUGCCUGGAAACCAGAUAACACAUUUUCCUAAGUCACGUCCACUUAAAUGGA-AAGAUUAUUAAUAAGAACUGUGUGUCUGGUGGAGAGGUAUAUUAUUUUAUAAUAGAAUAGU -......(((((....((((((((.((((..(((.((((((...((((......))))-..)))).....)).))).))))))))))))....))))).(((((((((...))))))))) ( -27.70) >DroSim_CAF1 5949 118 - 1 -UUUGAUUGCCUGGAAACCAGAUAACACAUUUUCCUAAGUCACGUCCACUUAAAUGGA-AAGAUGAUUAAUAAGAACUGUGUGUCUGGUGGAGAGGUCUAUUAUUUUAUAAUAGAAUAGU -........(((....((((((((.((((((((..(.(((((..((((......))))-....))))).)..)))).))))))))))))....)))((((((((...))))))))..... ( -29.50) >DroEre_CAF1 5863 120 - 1 UUCUGAUUAUCUCGAAACCAGAUUACACAUUUUCCUAUGUCACGUCCAGUUAAAUGGAAAAGAUGAUAAAUUAGAACUGUGUUUCUGGUGGAGAUGUCUUUUAUGUUACAUUAGAAUAGU (((((((((((((...((((((..(((((((((....((((((.((((......))))...).)))))....)))).))))).)))))).)))))).............))))))).... ( -31.41) >DroYak_CAF1 5954 119 - 1 -UCUGAUUACCUCGAAACCAGAUUACACAUUUUACUAAGUCACGUCCAGUUUAAUGGAAAAGAUGAUAAAUUAGAACUGUGUGUCUGGUGGAGAGGUCUCCUAUGUUAUAAUAGAAUAGU -.......(((((...(((((((.((((((((((....(((((.((((......))))...).))))....))))).))))))))))))...)))))...((((......))))...... ( -34.80) >consensus _UUUGAUUACCUGGAAACCAGAUAACACAUUUUCCUAAGUCACGUCCACUUAAAUGGA_AAGAUGAUUAAUAAGAACUGUGUGUCUGGUGGAGAGGUCUAUUAUUUUAUAAUAGAAUAGU ........((((....(((((((.(((((((((..(.((((((.((((......))))...).))))).)..)))).))))))))))))....))))...((((((((...)))))))). (-24.82 = -25.06 + 0.24)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:55:27 2006