
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 20,938,313 – 20,938,459 |
| Length | 146 |
| Max. P | 0.939211 |

| Location | 20,938,313 – 20,938,419 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 88.11 |
| Mean single sequence MFE | -25.48 |
| Consensus MFE | -18.82 |
| Energy contribution | -18.58 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.33 |
| SVM RNA-class probability | 0.690632 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 20938313 106 - 27905053 UUGCAUAGCUUAGUAAGGAACGCUAGCGCAUUAGUGGGGGUACUUCUUUCCCCACCAAAAAGUGCUCAAUAAUGCAAAAUGGACAUAUGUACAAAUAUGGAAAAGC ((((((.(((.(((.......))))))(((((.(((((((........))))))).....)))))......))))))......(((((......)))))....... ( -28.60) >DroSec_CAF1 1487 105 - 1 UGGCAUAGGUUAGUAAGGAAGACUAACGUAUUAAUGGGGGUACUUCUUCCCCCACCAA-AAGUGCUCAAUAAUGCAAAAUGGACAUAUGUACAAAUAUGGAAAAGC ....(((.((((((.......)))))).)))...((((((........))))))(((.-...(((........)))...))).(((((......)))))....... ( -27.50) >DroSim_CAF1 1487 106 - 1 UGGCAUAGGUUAGUAAGGAACACUAACGUUUUAAUGGGGGCACUUCUUCCCCCACCAAAAAGUGCUCAAUAAUGCAAAAUGGACAUAUGUACAAAUAUGGAAAAGC ........((((((.......))))))(((((..((((((........))))))(((.....(((........)))...))).(((((......)))))..))))) ( -24.70) >DroEre_CAF1 1344 106 - 1 GGGCGUCGCUUAGUAACAAACACUAACAUAUUAGUCGGGGCACUUCUUCCCCCACCAGAAAGUGCUCCGAAAUGCAAAAUGGACAUAUGUACAAAUAUGGAAAAGC ..((.(((((((((.......)))))........(((((((((((((.........)).)))))))))))...))........(((((......)))))))...)) ( -25.40) >DroYak_CAF1 1483 106 - 1 GGGCAUCGCUUAGUAACAAACACUAACAUAAUAGUCGGGGUACUUGCUCCCCCACCGAAAAGUGCUCCAAAAUGCAAAAUGGACAUAUGUACAAAUAUGGAAAAGC ..((((.(.(((((.......)))))).........(((((((((..((.......)).)))))))))...))))........(((((......)))))....... ( -21.20) >consensus UGGCAUAGCUUAGUAAGGAACACUAACGUAUUAGUGGGGGUACUUCUUCCCCCACCAAAAAGUGCUCAAUAAUGCAAAAUGGACAUAUGUACAAAUAUGGAAAAGC .......(.(((((.......)))))).......((((((........))))))(((.....(((........)))...))).(((((......)))))....... (-18.82 = -18.58 + -0.24)



| Location | 20,938,339 – 20,938,459 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 83.42 |
| Mean single sequence MFE | -33.78 |
| Consensus MFE | -23.18 |
| Energy contribution | -24.58 |
| Covariance contribution | 1.40 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.42 |
| Structure conservation index | 0.69 |
| SVM decision value | 1.22 |
| SVM RNA-class probability | 0.932426 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 20938339 120 + 27905053 AUUUUGCAUUAUUGAGCACUUUUUGGUGGGGAAAGAAGUACCCCCACUAAUGCGCUAGCGUUCCUUACUAAGCUAUGCAAAAAUAAAGUACUCCAAAGCAUUUUCGUUCGCCAUAGCCCA ....(((......(((.((((((((((((((..........)))))))))((((.((((............)))))))).....))))).)))....))).................... ( -27.90) >DroSec_CAF1 1513 118 + 1 AUUUUGCAUUAUUGAGCACUU-UUGGUGGGGGAAGAAGUACCCCCAUUAAUACGUUAGUCUUCCUUACUAACCUAUGCCAAA-AAAAGUGCUUCAAGGCAUUUUCGUUCGCCAUAGUCCA ...........((((((((((-((((((((((........)))))))).(((.((((((.......)))))).)))......-)))))))).))))(((..........)))........ ( -34.00) >DroSim_CAF1 1513 120 + 1 AUUUUGCAUUAUUGAGCACUUUUUGGUGGGGGAAGAAGUGCCCCCAUUAAAACGUUAGUGUUCCUUACUAACCUAUGCCAAAAAAAAGUGCUCCAAGGCAUUUUUGUUCGCCAUAGUCCA ....((.(((((.(((((((((((((((((((........)))))))).....(((((((.....)))))))..........)))))))))))...(((..........)))))))).)) ( -38.20) >DroEre_CAF1 1370 120 + 1 AUUUUGCAUUUCGGAGCACUUUCUGGUGGGGGAAGAAGUGCCCCGACUAAUAUGUUAGUGUUUGUUACUAAGCGACGCCCAAAUAAAGUGCUCCAUAUUAGUUUUAUUCGCCAUAGUCCA ............((((((((((.((((.((((........)))).))))........(((((.(((....))))))))......)))))))))).......................... ( -32.90) >DroYak_CAF1 1509 120 + 1 AUUUUGCAUUUUGGAGCACUUUUCGGUGGGGGAGCAAGUACCCCGACUAUUAUGUUAGUGUUUGUUACUAAGCGAUGCCCAAACAAAGUGCUCCAAAUAAGUUUUAUGCGCCAUAUUCCA .....(((((((((((((((((..(((.((((........)))).)))......((((((.....)))))).............)))))))))))))........))))........... ( -35.90) >consensus AUUUUGCAUUAUUGAGCACUUUUUGGUGGGGGAAGAAGUACCCCCACUAAUACGUUAGUGUUCCUUACUAAGCUAUGCCAAAAAAAAGUGCUCCAAAGCAUUUUUGUUCGCCAUAGUCCA .............(((((((((.(((((((((........))))))))).....((((((.....)))))).............)))))))))........................... (-23.18 = -24.58 + 1.40)

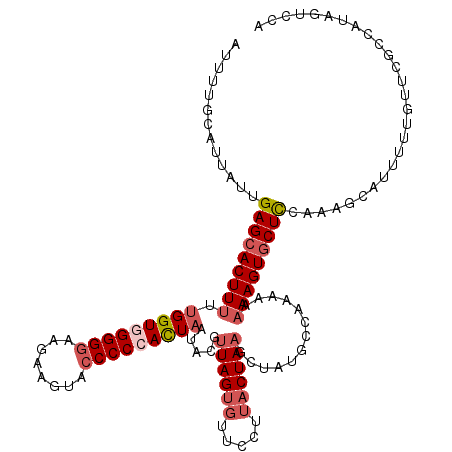
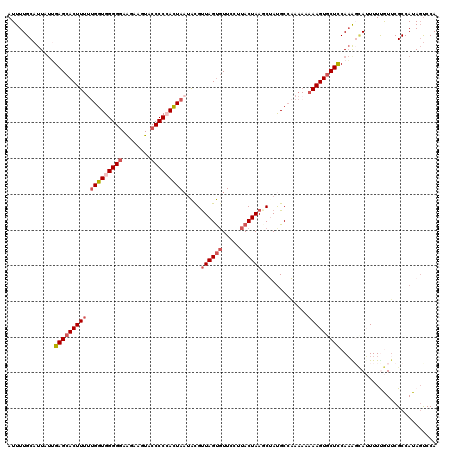
| Location | 20,938,339 – 20,938,459 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.42 |
| Mean single sequence MFE | -34.00 |
| Consensus MFE | -23.10 |
| Energy contribution | -23.78 |
| Covariance contribution | 0.68 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.50 |
| Structure conservation index | 0.68 |
| SVM decision value | 1.30 |
| SVM RNA-class probability | 0.939211 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 20938339 120 - 27905053 UGGGCUAUGGCGAACGAAAAUGCUUUGGAGUACUUUAUUUUUGCAUAGCUUAGUAAGGAACGCUAGCGCAUUAGUGGGGGUACUUCUUUCCCCACCAAAAAGUGCUCAAUAAUGCAAAAU ...((....)).........(((....(((((((((.....(((.((((............))))..)))...(((((((........)))))))...)))))))))......))).... ( -33.80) >DroSec_CAF1 1513 118 - 1 UGGACUAUGGCGAACGAAAAUGCCUUGAAGCACUUUU-UUUGGCAUAGGUUAGUAAGGAAGACUAACGUAUUAAUGGGGGUACUUCUUCCCCCACCAA-AAGUGCUCAAUAAUGCAAAAU ........((((........))))((((.(((((((.-..(((.(((.((((((.......)))))).)))...((((((........))))))))))-))))))))))........... ( -34.70) >DroSim_CAF1 1513 120 - 1 UGGACUAUGGCGAACAAAAAUGCCUUGGAGCACUUUUUUUUGGCAUAGGUUAGUAAGGAACACUAACGUUUUAAUGGGGGCACUUCUUCCCCCACCAAAAAGUGCUCAAUAAUGCAAAAU ........((((........))))...((((((((((...(((.....((((((.......)))))).......((((((........)))))))))))))))))))............. ( -35.00) >DroEre_CAF1 1370 120 - 1 UGGACUAUGGCGAAUAAAACUAAUAUGGAGCACUUUAUUUGGGCGUCGCUUAGUAACAAACACUAACAUAUUAGUCGGGGCACUUCUUCCCCCACCAGAAAGUGCUCCGAAAUGCAAAAU .........(((.......((((((((((((.((......)))).))..(((((.......)))))))))))))(((((((((((((.........)).)))))))))))..)))..... ( -31.80) >DroYak_CAF1 1509 120 - 1 UGGAAUAUGGCGCAUAAAACUUAUUUGGAGCACUUUGUUUGGGCAUCGCUUAGUAACAAACACUAACAUAAUAGUCGGGGUACUUGCUCCCCCACCGAAAAGUGCUCCAAAAUGCAAAAU ...........((((........(((((((((((((...((((....(.(((((.......)))))).........(((((....)))))))))....)))))))))))))))))..... ( -34.70) >consensus UGGACUAUGGCGAACAAAAAUGCUUUGGAGCACUUUAUUUUGGCAUAGCUUAGUAAGGAACACUAACGUAUUAGUGGGGGUACUUCUUCCCCCACCAAAAAGUGCUCAAUAAUGCAAAAU .........(((..((((.....))))(((((((((...........(.(((((.......))))))......(((((((........)))))))...))))))))).....)))..... (-23.10 = -23.78 + 0.68)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:55:23 2006