| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 20,931,357 – 20,931,470 |
| Length | 113 |
| Max. P | 0.940272 |

| Location | 20,931,357 – 20,931,470 |
|---|---|
| Length | 113 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.38 |
| Mean single sequence MFE | -28.57 |
| Consensus MFE | -15.08 |
| Energy contribution | -15.92 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.28 |
| Mean z-score | -2.51 |
| Structure conservation index | 0.53 |
| SVM decision value | 1.29 |
| SVM RNA-class probability | 0.940272 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 20931357 113 + 27905053 GAAUAUUUAGGCAUAGGGUUUUGUCCUUAUGAGGGCAAUUAUCCAACGCAUUCAUAGUCACUGGAAGAUAUCCCAAUCCUUUGAUGCUUCUAGUUU----CUAACCCUUU---AAUACUA ...((((.(((....((((.(((((((....)))))))..))))..........(((..((((((((.((((..........))))))))))))..----)))..)))..---))))... ( -28.30) >DroSec_CAF1 14859 113 + 1 GAAUAUUUAGGCAUAGGGUUUUGUCCUUAUGAGGGCAAUUAUCCAGCGCAUUCAUAGUAACUGGAAGAUAUCCCAAUCCUUUGAUGCUUCUAGUUU----CUAAGCCUUU---AAUACUA ...((((.((((...((((.(((((((....)))))))..))))..........(((.(((((((((.((((..........))))))))))))).----))).))))..---))))... ( -33.80) >DroSim_CAF1 23222 113 + 1 GAAUAUUUAGGCAUAGGGUUUUGUCCUUAUGAGGGCAAUUAUCCAGCGCAUUCAUAGUCACUGGAAGAUAUCCCAAUCCCUUGAUGCUUCUAGUUU----CUAAGCCUUU---AUUCCUA (((((...((((...((((.(((((((....)))))))..))))..........(((..((((((((.((((..........))))))))))))..----))).)))).)---))))... ( -35.80) >DroEre_CAF1 23220 113 + 1 GAAUAUUUAGGCAUAGGGUUUUGUCCUUAUGAGGACAAUUAUCCAGCGCAUUCAUAGAAACUGGUAAAUAUCCCAAUCCUUUGAUGCUUCUACUAG----CCAAGCCUUU---CAUACUU (((......(((...((((.(((((((....)))))))..))))...((((.((.((....(((........)))...)).))))))........)----))......))---)...... ( -26.40) >DroYak_CAF1 21558 113 + 1 GAAUAUUUAGGCAUAGGGUUUUGUCCUUAUGUGGACAAUUAUCCAGCGCAUUCAUAGAAACUGGAAAAUAUCCCAAUCCUUUGAUGCUUUUAGUUU----CUUGGCCUUU---AAUACUU ...((((.((((...((((.((((((......))))))..))))........((.(((((((((((..((((..........)))).)))))))))----))))))))..---))))... ( -29.60) >DroPer_CAF1 63950 120 + 1 GCAUUUUAAAGUACAAGGGUUUCUCAUUAUUUGGAAAUUUCUUCAACUUAGUUAUCGAAGCUGUAUAGUCUCUAAGGUUUUCUAUGCUUUUAAUUGAAACUUUGGAUUUUUCUGACUCCU ..........(((((...(((((..((((.(((((......)))))..))))....))))))))))(((((((((((((((..((.......)).))))))))))).......))))... ( -17.51) >consensus GAAUAUUUAGGCAUAGGGUUUUGUCCUUAUGAGGACAAUUAUCCAGCGCAUUCAUAGAAACUGGAAAAUAUCCCAAUCCUUUGAUGCUUCUAGUUU____CUAAGCCUUU___AAUACUA ........((((...((((.((((((......))))))..))))...(((.(((.((....(((........)))...)).)))))).................))))............ (-15.08 = -15.92 + 0.84)

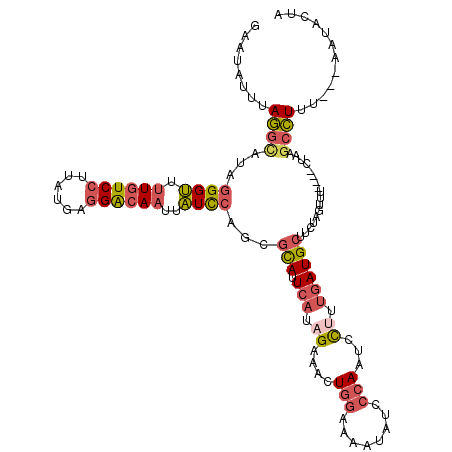

| Location | 20,931,357 – 20,931,470 |
|---|---|
| Length | 113 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.38 |
| Mean single sequence MFE | -26.83 |
| Consensus MFE | -12.72 |
| Energy contribution | -14.28 |
| Covariance contribution | 1.56 |
| Combinations/Pair | 1.34 |
| Mean z-score | -2.17 |
| Structure conservation index | 0.47 |
| SVM decision value | 0.31 |
| SVM RNA-class probability | 0.682380 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 20931357 113 - 27905053 UAGUAUU---AAAGGGUUAG----AAACUAGAAGCAUCAAAGGAUUGGGAUAUCUUCCAGUGACUAUGAAUGCGUUGGAUAAUUGCCCUCAUAAGGACAAAACCCUAUGCCUAAAUAUUC ..((((.---..((((((..----...((((..((((((.((.(((((((....)))))))..)).)).)))).))))....(((.(((....))).))))))))))))).......... ( -26.10) >DroSec_CAF1 14859 113 - 1 UAGUAUU---AAAGGCUUAG----AAACUAGAAGCAUCAAAGGAUUGGGAUAUCUUCCAGUUACUAUGAAUGCGCUGGAUAAUUGCCCUCAUAAGGACAAAACCCUAUGCCUAAAUAUUC .((((((---..((((.(((----...((((..((((((.((((((((((....)))))))).)).)).)))).))))....(((.(((....))).)))....))).)))).)))))). ( -29.00) >DroSim_CAF1 23222 113 - 1 UAGGAAU---AAAGGCUUAG----AAACUAGAAGCAUCAAGGGAUUGGGAUAUCUUCCAGUGACUAUGAAUGCGCUGGAUAAUUGCCCUCAUAAGGACAAAACCCUAUGCCUAAAUAUUC ...((((---(.((((.(((----...((((..((((((((..(((((((....)))))))..)).)).)))).))))....(((.(((....))).)))....))).))))...))))) ( -29.70) >DroEre_CAF1 23220 113 - 1 AAGUAUG---AAAGGCUUGG----CUAGUAGAAGCAUCAAAGGAUUGGGAUAUUUACCAGUUUCUAUGAAUGCGCUGGAUAAUUGUCCUCAUAAGGACAAAACCCUAUGCCUAAAUAUUC .(((((.---..((((..((----(((((....((((((.((((((((........)))).)))).)).)))))))))....(((((((....)))))))...))...))))..))))). ( -33.00) >DroYak_CAF1 21558 113 - 1 AAGUAUU---AAAGGCCAAG----AAACUAAAAGCAUCAAAGGAUUGGGAUAUUUUCCAGUUUCUAUGAAUGCGCUGGAUAAUUGUCCACAUAAGGACAAAACCCUAUGCCUAAAUAUUC .((((((---..((((..((----...(((...((((((.((((((((((....)))))).)))).)).))))..)))....((((((......))))))....))..)))).)))))). ( -29.10) >DroPer_CAF1 63950 120 - 1 AGGAGUCAGAAAAAUCCAAAGUUUCAAUUAAAAGCAUAGAAAACCUUAGAGACUAUACAGCUUCGAUAACUAAGUUGAAGAAAUUUCCAAAUAAUGAGAAACCCUUGUACUUUAAAAUGC .(((..........)))..((((((.......................)))))).((((((((((((......)))))))...((((((.....)).))))...)))))........... ( -14.10) >consensus AAGUAUU___AAAGGCUUAG____AAACUAGAAGCAUCAAAGGAUUGGGAUAUCUUCCAGUUACUAUGAAUGCGCUGGAUAAUUGCCCUCAUAAGGACAAAACCCUAUGCCUAAAUAUUC ............((((.(((.......((((..((((((.((((((((((....)))))))).)).))).))).))))....(((.((......)).)))....))).))))........ (-12.72 = -14.28 + 1.56)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:55:20 2006