
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 20,918,360 – 20,918,456 |
| Length | 96 |
| Max. P | 0.771240 |

| Location | 20,918,360 – 20,918,456 |
|---|---|
| Length | 96 |
| Sequences | 4 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 89.77 |
| Mean single sequence MFE | -22.78 |
| Consensus MFE | -17.42 |
| Energy contribution | -17.54 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.579203 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
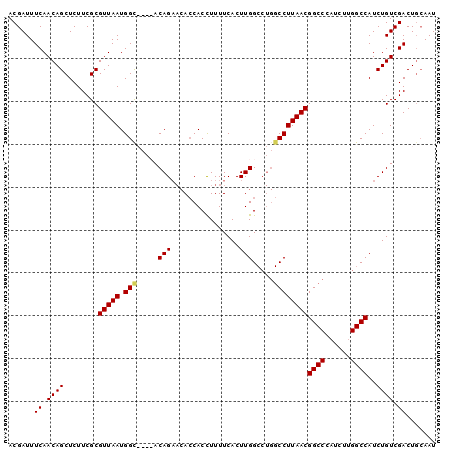
>3R_DroMel_CAF1 20918360 96 + 27905053 ACGAUUUCAACAGCUCUUCGCGUUAAUGGC----ACAGAACACCACCUUUUCACUUGGCCUGGCCUUAACGGCCCAUCUUGGCCAUCUGUCGACUGCAAU ............((...(((((((((.(((----.(((..((.............))..)))))))))))((((......))))....).)))..))... ( -24.22) >DroSec_CAF1 1692 100 + 1 ACGAUUUCAACAGCUCUUCGCGUUAAUGGUACACACAGAACACCACCUUUUCACUUGGCCUGGCCUUAACGGCCCAUCUUGGCCAUCUGUCGACUGCAAU ............((...(((......((((...........))))......((..(((((.((((.....))))......)))))..)).)))..))... ( -20.60) >DroSim_CAF1 10104 100 + 1 ACGAUUUCAACAGCUCUUCGCGUUAAUGGCACACACAGAACACCACCUUUUCACUUGGCCUGGCCUUAACGGCCCAUCUUGGCCAUCUGUCGACUGCAAU ........(((.((.....)))))....(((..((((((................(((((.((((.....))))......)))))))))).)..)))... ( -20.79) >DroEre_CAF1 9968 88 + 1 ACGAUUUCAACAGCUCUUCGCGUUAAUGGC----ACAG---AACACUUUUUCACUUGG-----CCUUAACGGCCGAUCUCGGCCAUCUGUCGACUCGAAU .(((..((.((((........(((((.(((----.(((---((......)))...)))-----)))))))(((((....)))))..)))).)).)))... ( -25.50) >consensus ACGAUUUCAACAGCUCUUCGCGUUAAUGGC____ACAGAACACCACCUUUUCACUUGGCCUGGCCUUAACGGCCCAUCUUGGCCAUCUGUCGACUGCAAU ......((.((((........(((((.(((.....(((................))).....))))))))((((......))))..)))).))....... (-17.42 = -17.54 + 0.12)


| Location | 20,918,360 – 20,918,456 |
|---|---|
| Length | 96 |
| Sequences | 4 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 89.77 |
| Mean single sequence MFE | -31.45 |
| Consensus MFE | -23.51 |
| Energy contribution | -24.70 |
| Covariance contribution | 1.19 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.53 |
| SVM RNA-class probability | 0.771240 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
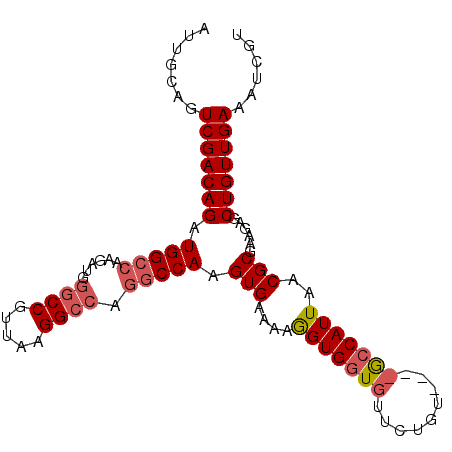
>3R_DroMel_CAF1 20918360 96 - 27905053 AUUGCAGUCGACAGAUGGCCAAGAUGGGCCGUUAAGGCCAGGCCAAGUGAAAAGGUGGUGUUCUGU----GCCAUUAACGCGAAGAGCUGUUGAAAUCGU .......(((((((..((((......))))(((((((((((((((..(....)..))))...))).----))).)))))........)))))))...... ( -33.10) >DroSec_CAF1 1692 100 - 1 AUUGCAGUCGACAGAUGGCCAAGAUGGGCCGUUAAGGCCAGGCCAAGUGAAAAGGUGGUGUUCUGUGUGUACCAUUAACGCGAAGAGCUGUUGAAAUCGU .......(((((((.(((((......((((.....)))).))))).(((....(((((((.........)))))))..)))......)))))))...... ( -31.00) >DroSim_CAF1 10104 100 - 1 AUUGCAGUCGACAGAUGGCCAAGAUGGGCCGUUAAGGCCAGGCCAAGUGAAAAGGUGGUGUUCUGUGUGUGCCAUUAACGCGAAGAGCUGUUGAAAUCGU .......(((((((.(((((......((((.....)))).))))).(((....(((((..(.......)..)))))..)))......)))))))...... ( -32.20) >DroEre_CAF1 9968 88 - 1 AUUCGAGUCGACAGAUGGCCGAGAUCGGCCGUUAAGG-----CCAAGUGAAAAAGUGUU---CUGU----GCCAUUAACGCGAAGAGCUGUUGAAAUCGU ...(((.(((((((..(((((....)))))(((((((-----(((...(((......))---))).----))).)))))........)))))))..))). ( -29.50) >consensus AUUGCAGUCGACAGAUGGCCAAGAUGGGCCGUUAAGGCCAGGCCAAGUGAAAAGGUGGUGUUCUGU____GCCAUUAACGCGAAGAGCUGUUGAAAUCGU .......(((((((.(((((......((((.....)))).))))).(((....(((((((.........)))))))..)))......)))))))...... (-23.51 = -24.70 + 1.19)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:55:07 2006