
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 20,873,298 – 20,873,625 |
| Length | 327 |
| Max. P | 0.836983 |

| Location | 20,873,298 – 20,873,404 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.00 |
| Mean single sequence MFE | -27.20 |
| Consensus MFE | -18.01 |
| Energy contribution | -17.81 |
| Covariance contribution | -0.20 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.59 |
| SVM RNA-class probability | 0.793060 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
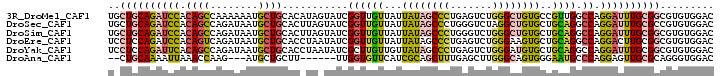
>3R_DroMel_CAF1 20873298 106 + 27905053 -------------GCCU-AUCUACCAGAAUAUUAGCCAAGGUACACAUUUGUACCUGCAAACGUACCUAGGUACGAAAGCUGUCACCUUGCGCAUCACUGGCGACGAUACAUUUCGCCGA -------------....-................((((((((((((.((((((((((..........)))))))))).).))).)))))).)).....((((((.........)))))). ( -31.00) >DroSec_CAF1 2514 109 + 1 AAAGU----UUAUUCCU-AUUUACCAAAAUAUUACCCUAGGUACAGAUUUGUACCUG------UAUCUAGGUACGAAAGCUGCAACCUUGCACAUCACUGGCGGCGAUACAUUUCGCAGA .....----........-.....(((.........(((((((((((........)))------)))))))).......(.((((....))))).....)))..((((......))))... ( -26.50) >DroSim_CAF1 1192 115 + 1 AAAGU----UUAUUCCU-AUUUACCAAAAUAUUAGCCUAGGUACAGAUUUGUACCUGCAAACGUACCUAGGUUCGAAAGCUGUCACCUUGCGCAUCACUGGCGACGAUACAUUUCGCAGA .....----........-...............(((((((((((.(.(((((....)))))))))))))))))(((((..((((...((((.((....)))))).))))..))))).... ( -27.70) >DroEre_CAF1 1275 114 + 1 AAAGU-----UUUUCCUUUGCCACCA-AAUAUCAACCUAGGUACAUGUGUGUACCUGCUACCUUACCUAGGUACGAAAGCUGUCACCUUUCGCAUCACUGGCGACGAUACAUUUUGCCGA ...((-----...((..((((((...-.......(((((((((...(((.((....)))))..))))))))).((((((.......))))))......)))))).)).)).......... ( -26.40) >DroYak_CAF1 1190 118 + 1 -AAGUUCGUUUGUUUCUUUCCCACCAAAAUAUCAUCAUAGGUACACACUUGUACCUAC-GCCUUACCUAGGUACGAAAGCUGUCACCUUACGCAUCACUGGCGACGAUACAUUUCGCCGA -...(((((............................((((((((....)))))))).-((((.....))))))))).((.((......)))).....((((((.........)))))). ( -24.40) >consensus AAAGU____UUAUUCCU_AUCUACCAAAAUAUUAGCCUAGGUACACAUUUGUACCUGC_AACGUACCUAGGUACGAAAGCUGUCACCUUGCGCAUCACUGGCGACGAUACAUUUCGCCGA ......................(((............((((((((....))))))))............))).(((((..((((.((............)).)))).....))))).... (-18.01 = -17.81 + -0.20)

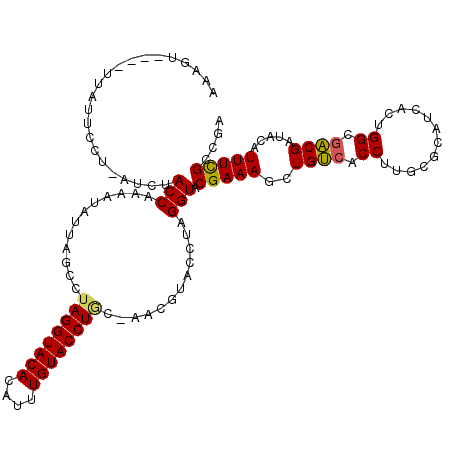
| Location | 20,873,298 – 20,873,404 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.00 |
| Mean single sequence MFE | -33.50 |
| Consensus MFE | -23.24 |
| Energy contribution | -23.76 |
| Covariance contribution | 0.52 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.71 |
| SVM RNA-class probability | 0.830390 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 20873298 106 - 27905053 UCGGCGAAAUGUAUCGUCGCCAGUGAUGCGCAAGGUGACAGCUUUCGUACCUAGGUACGUUUGCAGGUACAAAUGUGUACCUUGGCUAAUAUUCUGGUAGAU-AGGC------------- ..(((((......)))))(((((....(((((((....)......(((((....))))).))))(((((((....)))))))..)).......)))))....-....------------- ( -33.90) >DroSec_CAF1 2514 109 - 1 UCUGCGAAAUGUAUCGCCGCCAGUGAUGUGCAAGGUUGCAGCUUUCGUACCUAGAUA------CAGGUACAAAUCUGUACCUAGGGUAAUAUUUUGGUAAAU-AGGAAUAA----ACUUU ..(((.(((.((((..((((((....)).)).((((.((((.(((.((((((.....------.))))))))).))))))))..))..))))))).)))...-........----..... ( -30.20) >DroSim_CAF1 1192 115 - 1 UCUGCGAAAUGUAUCGUCGCCAGUGAUGCGCAAGGUGACAGCUUUCGAACCUAGGUACGUUUGCAGGUACAAAUCUGUACCUAGGCUAAUAUUUUGGUAAAU-AGGAAUAA----ACUUU (((((.(((((((((...((((....)).)).((((............)))).))))))))))))))((((....))))((((.(((((....)))))...)-))).....----..... ( -31.70) >DroEre_CAF1 1275 114 - 1 UCGGCAAAAUGUAUCGUCGCCAGUGAUGCGAAAGGUGACAGCUUUCGUACCUAGGUAAGGUAGCAGGUACACACAUGUACCUAGGUUGAUAUU-UGGUGGCAAAGGAAAA-----ACUUU ...............((((((((....(((((((.......)))))))(((((((((..((.(........)))...)))))))))......)-)))))))((((.....-----.)))) ( -32.20) >DroYak_CAF1 1190 118 - 1 UCGGCGAAAUGUAUCGUCGCCAGUGAUGCGUAAGGUGACAGCUUUCGUACCUAGGUAAGGC-GUAGGUACAAGUGUGUACCUAUGAUGAUAUUUUGGUGGGAAAGAAACAAACGAACUU- ..(((((.........)))))(((....(((..(....)..(((((.((((.(((((...(-(((((((((....))))))))))....))))).)))).)))))......))).))).- ( -39.50) >consensus UCGGCGAAAUGUAUCGUCGCCAGUGAUGCGCAAGGUGACAGCUUUCGUACCUAGGUAAGGU_GCAGGUACAAAUCUGUACCUAGGCUAAUAUUUUGGUAGAU_AGGAAUAA____ACUUU ...((((((.((...((((((............)))))).))))))))(((.(((((.......(((((((....))))))).......))))).)))...................... (-23.24 = -23.76 + 0.52)

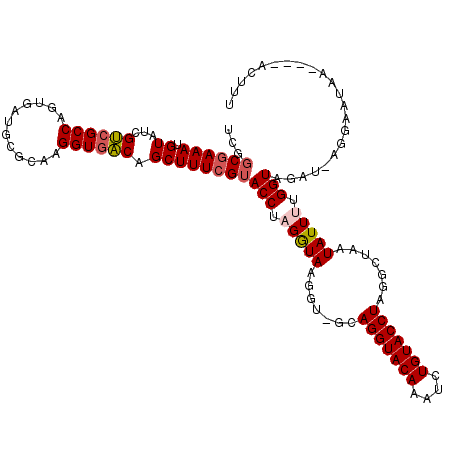

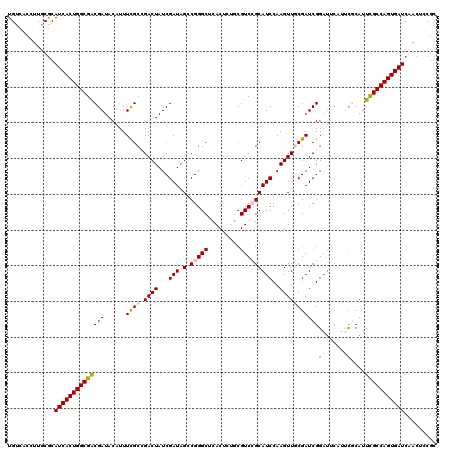
| Location | 20,873,364 – 20,873,484 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.25 |
| Mean single sequence MFE | -38.34 |
| Consensus MFE | -26.46 |
| Energy contribution | -26.94 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.546112 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 20873364 120 + 27905053 UGUCACCUUGCGCAUCACUGGCGACGAUACAUUUCGCCGACUAUCGAUAGACGGGCUCACUCUGCGUCCGCAUCCAAGUUGCGAUCGGAUUCAUUCGCAUUCGCCAGUGAUCAACUCCGC ...........(.((((((((((((((......((((.((((...(((.(.(((((.(.....).)))))))))..)))))))))))((.....))....))))))))))))........ ( -36.20) >DroSec_CAF1 2583 120 + 1 UGCAACCUUGCACAUCACUGGCGGCGAUACAUUUCGCAGACUAUCGAUAGCCGGGCUCACUCUGCGUCCGCAUCCAAGUUGCGAUCGGAUUCAUUCGCAUUCGCCAGUGAUCAACUCCGC ((((....)))).((((((((((((((......)))).((((...(((.((.((((.(.....).)))))))))..))))((((..........))))...))))))))))......... ( -40.20) >DroSim_CAF1 1267 120 + 1 UGUCACCUUGCGCAUCACUGGCGACGAUACAUUUCGCAGACUAUCGAUAGCCGGGCUCACUCUGCGUCCGCAUCCAAGUUGCGAUCGGAUUCAUUCGCAUUCGCCAGUGAUCAACUCCGC ...........(.((((((((((((((......((((((.((...(((.((.((((.(.....).)))))))))..)))))))))))((.....))....))))))))))))........ ( -36.50) >DroEre_CAF1 1349 114 + 1 UGUCACCUUUCGCAUCACUGGCGACGAUACAUUUUGCCGACUAUCGAUAGUCGGGCUCACUGGCCGUCGGCAUCCAAGUUCCGAUCGG------AUUCAUUCGCCAGUGAUCCACUCCGC ...........(.(((((((((((..........(((((((...((.....))((((....))))))))))).......(((....))------).....))))))))))).)....... ( -40.60) >DroYak_CAF1 1268 120 + 1 UGUCACCUUACGCAUCACUGGCGACGAUACAUUUCGCCGACUAUCGAUAGCCGGGCUCACUGACCGUCGGCAUCCAAGUUCCGAUCGGAUUCGGAUUCAUUUGCCAGUGAUCCACUCCGC ...........(.(((((((((((...........((((((..(((..(((...)))...)))..))))))........(((((......))))).....))))))))))).)....... ( -38.20) >consensus UGUCACCUUGCGCAUCACUGGCGACGAUACAUUUCGCCGACUAUCGAUAGCCGGGCUCACUCUGCGUCCGCAUCCAAGUUGCGAUCGGAUUCAUUCGCAUUCGCCAGUGAUCAACUCCGC .............((((((((((((((......((((.((((...(((.(.(((((.........)))))))))..)))))))))))((.....))....)))))))))))......... (-26.46 = -26.94 + 0.48)

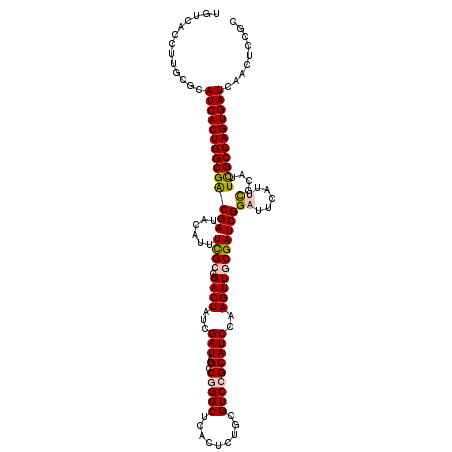
| Location | 20,873,364 – 20,873,484 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.25 |
| Mean single sequence MFE | -45.96 |
| Consensus MFE | -32.48 |
| Energy contribution | -33.32 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.32 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.54 |
| SVM RNA-class probability | 0.774832 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
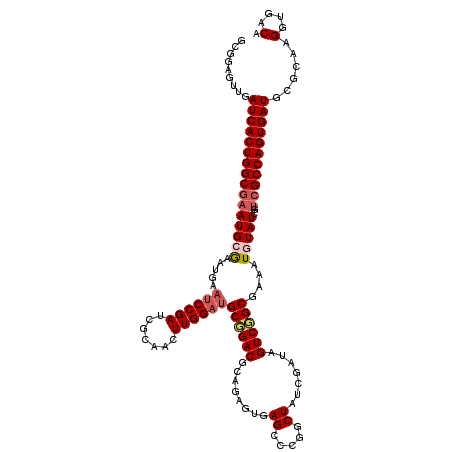
>3R_DroMel_CAF1 20873364 120 - 27905053 GCGGAGUUGAUCACUGGCGAAUGCGAAUGAAUCCGAUCGCAACUUGGAUGCGGACGCAGAGUGAGCCCGUCUAUCGAUAGUCGGCGAAAUGUAUCGUCGCCAGUGAUGCGCAAGGUGACA (((.(.....((((((((((.(((((.((....)).))))).....((((((..(((...))).(((.(.(((....))).))))....)))))).))))))))))).)))..(....). ( -41.90) >DroSec_CAF1 2583 120 - 1 GCGGAGUUGAUCACUGGCGAAUGCGAAUGAAUCCGAUCGCAACUUGGAUGCGGACGCAGAGUGAGCCCGGCUAUCGAUAGUCUGCGAAAUGUAUCGCCGCCAGUGAUGUGCAAGGUUGCA ((((..(((((((((((((..(((((.((....)).))))).....((((((..((((((.(((((...))).....)).))))))...))))))..))))))))))...)))..)))). ( -46.70) >DroSim_CAF1 1267 120 - 1 GCGGAGUUGAUCACUGGCGAAUGCGAAUGAAUCCGAUCGCAACUUGGAUGCGGACGCAGAGUGAGCCCGGCUAUCGAUAGUCUGCGAAAUGUAUCGUCGCCAGUGAUGCGCAAGGUGACA (((.(.....((((((((((.(((((.((....)).))))).....((((((..((((((.(((((...))).....)).))))))...)))))).))))))))))).)))..(....). ( -46.80) >DroEre_CAF1 1349 114 - 1 GCGGAGUGGAUCACUGGCGAAUGAAU------CCGAUCGGAACUUGGAUGCCGACGGCCAGUGAGCCCGACUAUCGAUAGUCGGCAAAAUGUAUCGUCGCCAGUGAUGCGAAAGGUGACA ......((.(((((((((((.(((((------((((.......))))))(((((((((......)))((.....))...))))))........)))))))))))))).))...(....). ( -46.30) >DroYak_CAF1 1268 120 - 1 GCGGAGUGGAUCACUGGCAAAUGAAUCCGAAUCCGAUCGGAACUUGGAUGCCGACGGUCAGUGAGCCCGGCUAUCGAUAGUCGGCGAAAUGUAUCGUCGCCAGUGAUGCGUAAGGUGACA .......(.(((((((((......((((((.(((....)))..))))))...((((((((....(((.(((((....))))))))....)).))))))))))))))).)....(....). ( -48.10) >consensus GCGGAGUUGAUCACUGGCGAAUGCGAAUGAAUCCGAUCGCAACUUGGAUGCGGACGCAGAGUGAGCCCGGCUAUCGAUAGUCGGCGAAAUGUAUCGUCGCCAGUGAUGCGCAAGGUGACA .........((((((((((((((((.....((((((.......))))))((((((........((.....)).......))))))....)))))..)))))))))))......(....). (-32.48 = -33.32 + 0.84)

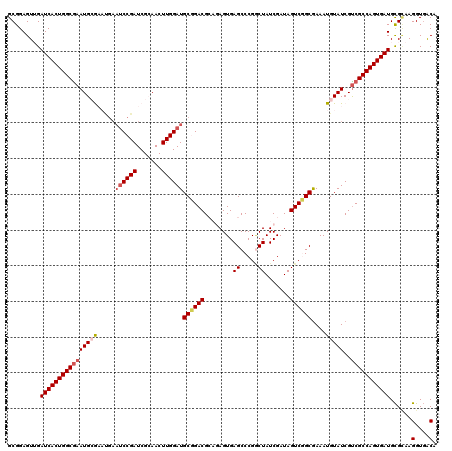
| Location | 20,873,484 – 20,873,585 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 79.74 |
| Mean single sequence MFE | -36.37 |
| Consensus MFE | -23.13 |
| Energy contribution | -23.75 |
| Covariance contribution | 0.62 |
| Combinations/Pair | 1.46 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.74 |
| SVM RNA-class probability | 0.836983 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 20873484 101 + 27905053 UGCUGCAGAUCCACAGCCAAAAAAUGCUGCACAUAGUAUCGGUUGUUAUUAUAGCCCUGAGUCUGGGCUGUGCCGUUGGCCAGGAUUUGCGGCGUGUGGAC .(((((((((((...(((((...((((((....))))))((((.......(((((((.......))))))))))))))))..)))))))))))........ ( -42.11) >DroSec_CAF1 2703 101 + 1 UGCUGCAGAUCCACAGCCAGAUAAUGCUGCACUUAGUAUCGGUUGUUAUUAUAGCCCUGGGUCUAGGCUGUGCUGCAGGCCAGGAUUUGCGCCGUGUGGAC .((.((((((((((((((((((.((((((....)))))).((((((....))))))....)))).))))))((.....))..))))))))))......... ( -35.80) >DroSim_CAF1 1387 101 + 1 UGCUGCAGAUCCACAGCCAGAUAAUGCUGCACUUAGUAUCGGUUGUUAUUAUAGCCCUGGGUCUGGGCUGUGCUGCAGGCCAGGAUUUGCGGCGUGUGGAC .(((((((((((....((((...((((((....)))))).((((((....))))))))))...(((.(((.....))).))))))))))))))........ ( -41.90) >DroEre_CAF1 1463 101 + 1 UCCUCCAGAUCCACAGUCAGAUAAUGCUGCACCUAAUAUCGGUUGUUAUUAUAGCCCUGAGUCUGGGAAGUGCUGCAGGCCAGGACUUGCGGCGUGUGGAC (((((((((..(((((.((.....)))))...........((((((....)))))).))..))))))....((((((((......))))))))....))). ( -32.20) >DroYak_CAF1 1388 101 + 1 UCCUCCAGAUUCACAGCCAGAUAAUGCUGCACCUAAUAUCGCUUGUUGUUAUAGCCCUGAGUCUGGGAUGUGCUGCAGGCCAGGAUUUGCGGCGUGUGGAC (((((((((((((..((..((((((...((..........))..))))))...))..))))))))))(..(((((((((......)))))))))..)))). ( -33.80) >DroAna_CAF1 2953 90 + 1 --CUGCAAAAUUAAACCAAG---AUGCUGCUU------UUGGUGUUCAUCGCAGCUUUGAGCUUGGGCAGUGGGAAUGCCCAGGAGUUGCGCAGGGUGGAC --((((........((((((---(......))------))))).......((((((.....((((((((.......))))))))))))))))))....... ( -32.40) >consensus UGCUGCAGAUCCACAGCCAGAUAAUGCUGCACCUAGUAUCGGUUGUUAUUAUAGCCCUGAGUCUGGGCUGUGCUGCAGGCCAGGAUUUGCGGCGUGUGGAC ..((((((((((.((((........))))...........((((((...((((((((.......))))))))..)))).)).))))))))))......... (-23.13 = -23.75 + 0.62)
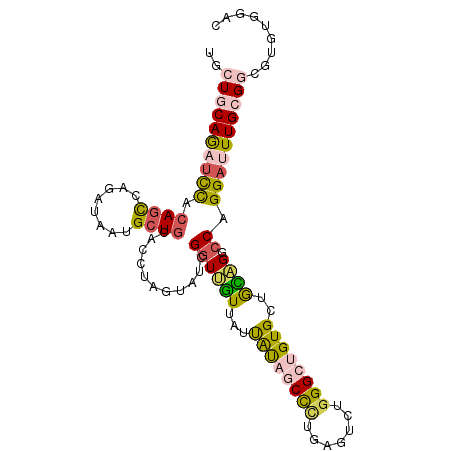

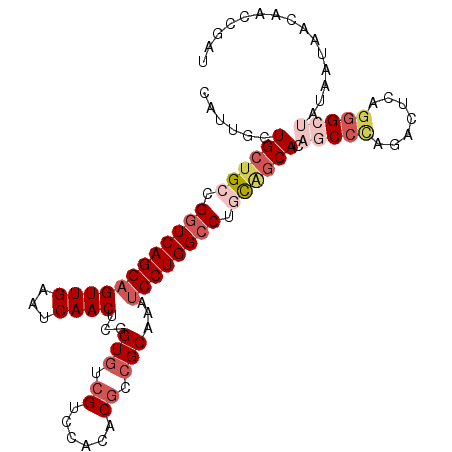
| Location | 20,873,521 – 20,873,625 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 87.50 |
| Mean single sequence MFE | -37.90 |
| Consensus MFE | -27.71 |
| Energy contribution | -29.68 |
| Covariance contribution | 1.97 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.93 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.552249 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 20873521 104 - 27905053 CAUUGCUGCUGCCGGUCAGGAGUUGAAUCAACUCGGUGUCGUCCACACGCCGCAAAUCCUGGCCAACGGCACAGCCCAGACUCAGGGCUAUAAUAACAACCGAU ......(((((..((((((((((((...)))).((((((.(....)))))))....))))))))..))))).(((((.......)))))............... ( -36.80) >DroSec_CAF1 2740 104 - 1 CAUUGCUGCUGCCGGUCAGGUGUUGAAUCAACUCCGUGUCGUCCACACGGCGCAAAUCCUGGCCUGCAGCACAGCCUAGACCCAGGGCUAUAAUAACAACCGAU ......((((((.(((((((.((((...))))...(((((((....)))))))....))))))).)))))).(((((.......)))))............... ( -39.40) >DroSim_CAF1 1424 104 - 1 CAUUGCUGCUGCCGGUCAGGAGUUGAAUCAACUCCGUAUCGUCCACACGCCGCAAAUCCUGGCCUGCAGCACAGCCCAGACCCAGGGCUAUAAUAACAACCGAU ...((((((.(((((...(((((((...)))))))((..(((....)))..)).....)))))..)))))).(((((.......)))))............... ( -37.60) >DroEre_CAF1 1500 104 - 1 CAUUGCUGCUGCCGGUCAGGAGUUGAAUCAACUCGGUGUCGUCCACACGCCGCAAGUCCUGGCCUGCAGCACUUCCCAGACUCAGGGCUAUAAUAACAACCGAU ......((((((.((((((((.(((........((((((.(....)))))))))).)))))))).))))))...(((.......)))................. ( -37.80) >DroYak_CAF1 1425 104 - 1 CAUUGCUGCUGCCGGUCAGGAGUUGAAUCAACUCGGUGUCGUCCACACGCCGCAAAUCCUGGCCUGCAGCACAUCCCAGACUCAGGGCUAUAACAACAAGCGAU .(((((((((((.((((((((((((...)))).((((((.(....)))))))....)))))))).)))))....(((.......)))...........)))))) ( -41.00) >DroAna_CAF1 2980 103 - 1 CGUUGUUGCUGCCGGUCAGGAGUUGAAUCAACUCGGUGUCGUCCACCCUGCGCAACUCCUGGGCAUUCCCACUGCCCAAGCUCAAAGCUGCGAUGAACACCAA- ((((((.(((...(((.((((((((...((....((((.....)))).))..))))))))(((((.......)))))..)))...))).))))))........- ( -34.80) >consensus CAUUGCUGCUGCCGGUCAGGAGUUGAAUCAACUCGGUGUCGUCCACACGCCGCAAAUCCUGGCCUGCAGCACAGCCCAGACUCAGGGCUAUAAUAACAACCGAU ......((((((.((((((((((((...))))...(((.((......)).)))...)))))))).)))))).(((((.......)))))............... (-27.71 = -29.68 + 1.97)

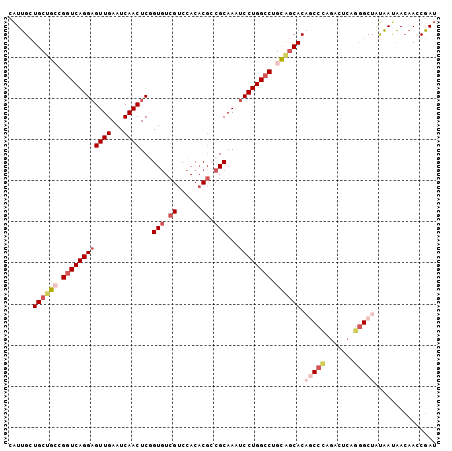
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:54:50 2006