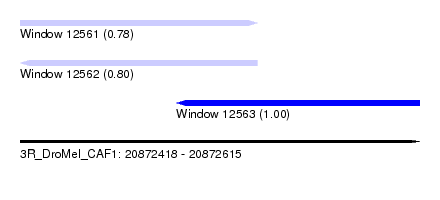
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 20,872,418 – 20,872,615 |
| Length | 197 |
| Max. P | 0.997885 |

| Location | 20,872,418 – 20,872,535 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.00 |
| Mean single sequence MFE | -29.22 |
| Consensus MFE | -20.42 |
| Energy contribution | -21.07 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.36 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.55 |
| SVM RNA-class probability | 0.779152 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 20872418 117 + 27905053 ---UUGAAUGCUAAACUUAUAAGAUUCUUUCGCAUUUAUUAGGCAACGAUAGUGGGGGCUACAGCUACAGCUCCCCGCCGAGCAACAGCUACUUGCCUCCAGCCACACCUGCUCCGGAGU ---.(((((((.(((............))).)))))))...(((.((....))((((((...((((...((((......))))...))))....)))))).)))...((......))... ( -31.30) >DroSec_CAF1 1622 117 + 1 ---UUAAAAGCUUAACUCAUAAGAUUUUUUUGCAUUUAUCAGGCAAUGAUAGUGGGGGCUACAGCUAUAGCUCCCCGCCGAGCAACAGCUACUUGCCUCCAGCCACUCCUGCUCCGGAGU ---..(((((((((.....)))).)))))..((..((((((.....))))))(((((((...((((...((((......))))...))))....))))))))).(((((......))))) ( -33.20) >DroSim_CAF1 324 117 + 1 ---UUAAAAGCUUAACUCAUAAGAUUAUUUUGAAUUUAUCAGGCAACGAUAGUGGGGGCUACAGCUACAGCUCCCCGCCGAGCAACAGCUACUUGCCUCCAGCCACUCCUGCUCCGGAGU ---((((((.((((.....))))....))))))........(((.((....))((((((...((((...((((......))))...))))....)))))).)))(((((......))))) ( -34.40) >DroEre_CAF1 310 120 + 1 AAGUUGAAAUCUAGACUUACUUGAUUCUUUUGCAUUUAUUAGGCAACGACAGUGGGGGCUACAGCUACAGCUCCCCACCGAGCAACAGCUACCUACCUCCAGCCACUCCUGCACCGGAGU .(((((.((((.((.....)).))))...((((........(....)..(.(((((((((........))).)))))).).)))))))))..............(((((......))))) ( -31.60) >DroYak_CAF1 336 120 + 1 AAGUUGAAAGCUAGACUUACUUGAUUCUUUUGCAUUUACUAGGCAACGACAGUGGGGGCUACAGCUACAGCUCCCCACCCAGCAACAGUUACCUACCUCCAGCCACUCCUGCUCCGGAGU .........(((.((..((..(((((...((((........(....)....(((((((((........))).))))))...)))).)))))..))..)).))).(((((......))))) ( -30.50) >DroAna_CAF1 1838 95 + 1 ------------------------UCCUUUUU-CUAAAUCAGGCAAUGAUAGCGGAGGAUAUAGCUAUAAUCCCCCACCAAAUAACAACUACCUGCCACCUGCAACACCUGCCCCGGAUU ------------------------........-...((((.((((.((.....((.((((.........)))).)).................(((.....))).))..))))...)))) ( -14.30) >consensus ___UUGAAAGCUAAACUUAUAAGAUUCUUUUGCAUUUAUCAGGCAACGAUAGUGGGGGCUACAGCUACAGCUCCCCACCGAGCAACAGCUACCUGCCUCCAGCCACUCCUGCUCCGGAGU .........................................(((.......(((((((((........))).))))))..(((....)))...........)))(((((......))))) (-20.42 = -21.07 + 0.64)


| Location | 20,872,418 – 20,872,535 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.00 |
| Mean single sequence MFE | -37.08 |
| Consensus MFE | -24.99 |
| Energy contribution | -24.68 |
| Covariance contribution | -0.30 |
| Combinations/Pair | 1.35 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.61 |
| SVM RNA-class probability | 0.798081 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 20872418 117 - 27905053 ACUCCGGAGCAGGUGUGGCUGGAGGCAAGUAGCUGUUGCUCGGCGGGGAGCUGUAGCUGUAGCCCCCACUAUCGUUGCCUAAUAAAUGCGAAAGAAUCUUAUAAGUUUAGCAUUCAA--- .((((((.((....))..))))))((.....))((.((((..((((((.((((......)))))))).((.((((............)))).))..........))..))))..)).--- ( -34.60) >DroSec_CAF1 1622 117 - 1 ACUCCGGAGCAGGAGUGGCUGGAGGCAAGUAGCUGUUGCUCGGCGGGGAGCUAUAGCUGUAGCCCCCACUAUCAUUGCCUGAUAAAUGCAAAAAAAUCUUAUGAGUUAAGCUUUUAA--- (((((......)))))((((..((((((...((((.....))))((((.(((((....))))))))).......))))))................((....))....)))).....--- ( -37.30) >DroSim_CAF1 324 117 - 1 ACUCCGGAGCAGGAGUGGCUGGAGGCAAGUAGCUGUUGCUCGGCGGGGAGCUGUAGCUGUAGCCCCCACUAUCGUUGCCUGAUAAAUUCAAAAUAAUCUUAUGAGUUAAGCUUUUAA--- (((((......)))))((((..((((((...((((.....))))((((.((((......)))))))).......))))))....((((((.((.....)).)))))).)))).....--- ( -38.00) >DroEre_CAF1 310 120 - 1 ACUCCGGUGCAGGAGUGGCUGGAGGUAGGUAGCUGUUGCUCGGUGGGGAGCUGUAGCUGUAGCCCCCACUGUCGUUGCCUAAUAAAUGCAAAAGAAUCAAGUAAGUCUAGAUUUCAACUU .(((((((.((....))))))))).((((((((.(.....((((((((.((((......)))))))))))).)))))))))............(((((.((.....)).)))))...... ( -41.00) >DroYak_CAF1 336 120 - 1 ACUCCGGAGCAGGAGUGGCUGGAGGUAGGUAACUGUUGCUGGGUGGGGAGCUGUAGCUGUAGCCCCCACUGUCGUUGCCUAGUAAAUGCAAAAGAAUCAAGUAAGUCUAGCUUUCAACUU (((((......)))))((((((((.((((((((....((..(((((((.((((......))))))))))))).)))))))).)...(((...........)))..)))))))........ ( -41.50) >DroAna_CAF1 1838 95 - 1 AAUCCGGGGCAGGUGUUGCAGGUGGCAGGUAGUUGUUAUUUGGUGGGGGAUUAUAGCUAUAUCCUCCGCUAUCAUUGCCUGAUUUAG-AAAAAGGA------------------------ ..((..((.(((((..((((((((((((....))))))))))(((((((((.........)))))))))...))..))))).))..)-).......------------------------ ( -30.10) >consensus ACUCCGGAGCAGGAGUGGCUGGAGGCAAGUAGCUGUUGCUCGGCGGGGAGCUGUAGCUGUAGCCCCCACUAUCGUUGCCUAAUAAAUGCAAAAGAAUCUUAUAAGUUUAGCUUUCAA___ (((((......)))))......((((((...((((.....))))((((.((((......)))))))).......))))))........................................ (-24.99 = -24.68 + -0.30)



| Location | 20,872,495 – 20,872,615 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.42 |
| Mean single sequence MFE | -42.96 |
| Consensus MFE | -40.08 |
| Energy contribution | -39.32 |
| Covariance contribution | -0.76 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.93 |
| SVM decision value | 2.95 |
| SVM RNA-class probability | 0.997885 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 20872495 120 - 27905053 AUAGUUGUUGUUAGGCGGUGGCAGGUAGUUGUUGUUCGGCGGCGGAUUGUACUGGUAUCCAUUGUUCGGCGGAGGCAGGUACUCCGGAGCAGGUGUGGCUGGAGGCAAGUAGCUGUUGCU ................((..((((.((.(((((.((((((.((((((.........)))).((((((..(((((.......)))))))))))..)).))))))))))).)).))))..)) ( -44.20) >DroSec_CAF1 1699 120 - 1 AUAGUUGUUGUUAGGCGGUGGCAGGUAGUUGUUGUUCGGUGGCGGAUUGUACUGGUAUCCAUUGUUCGGCGGAGGCAGGUACUCCGGAGCAGGAGUGGCUGGAGGCAAGUAGCUGUUGCU ................((..((((.((.(((((.((((((.((((((.........)))).((((((..(((((.......)))))))))))..)).))))))))))).)).))))..)) ( -42.90) >DroSim_CAF1 401 120 - 1 AUAGUUGUUGUUAGGCGGUGGCAGGUAGUUGUUGUUCGGCGGCGGAUUGUACUGGUAUCCAUUGUUCGGCGGAGGCAGGUACUCCGGAGCAGGAGUGGCUGGAGGCAAGUAGCUGUUGCU ................((..((((.((.(((((.((((((.((((((.........)))).((((((..(((((.......)))))))))))..)).))))))))))).)).))))..)) ( -44.40) >DroEre_CAF1 390 120 - 1 AUAGUUGUUGUUCGGCGGUGGCAGGUAGUUGUUGUUCGGCGGUGGAUUGUACUGGUAUCCAUUGUUCGGCGGAGGCAGGUACUCCGGUGCAGGAGUGGCUGGAGGUAGGUAGCUGUUGCU ...((((.....))))((..((((.((.((((((((..(((((((((.........)))))))))..)))))...(((.((((((......)))))).)))....))).)).))))..)) ( -42.70) >DroYak_CAF1 416 120 - 1 AUAGUUGUUGUUGGGCGGUGGCAGGUAGUUGUUGUUUGGCGGCGGAUUGUACUGGUAUCCAUUGUUCGGCGGAGGCAGGUACUCCGGAGCAGGAGUGGCUGGAGGUAGGUAACUGUUGCU (((((((((((..(((((..((.....))..)))))..)))))........(..((.(((..(((((..(((((.......)))))))))))))...))..)........)))))).... ( -40.60) >consensus AUAGUUGUUGUUAGGCGGUGGCAGGUAGUUGUUGUUCGGCGGCGGAUUGUACUGGUAUCCAUUGUUCGGCGGAGGCAGGUACUCCGGAGCAGGAGUGGCUGGAGGCAAGUAGCUGUUGCU ................((..((((.((.(((((.((((((.((((((.........)))).((((((..(((((.......)))))))))))..)).))))))))))).)).))))..)) (-40.08 = -39.32 + -0.76)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:54:45 2006