
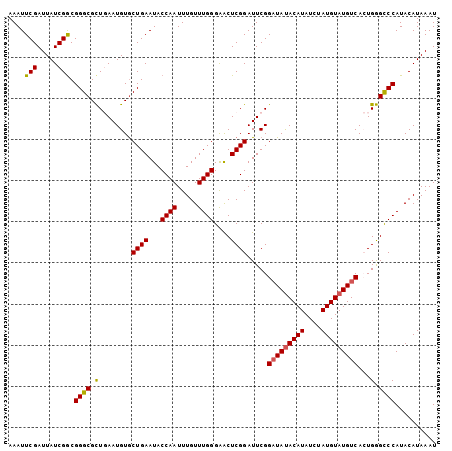
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 20,870,494 – 20,870,619 |
| Length | 125 |
| Max. P | 0.999522 |

| Location | 20,870,494 – 20,870,598 |
|---|---|
| Length | 104 |
| Sequences | 5 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 94.80 |
| Mean single sequence MFE | -27.08 |
| Consensus MFE | -24.04 |
| Energy contribution | -23.88 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.38 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.72 |
| SVM RNA-class probability | 0.832756 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 20870494 104 + 27905053 AAAUUCGAUUAUCGGCGGGCGCUGAAUGUGCUGAAUACCAAUUUGUUUGGGAACUCGGAUUCGGAUAUACAUAUCUAUGUAUGUCACUGGGCCCAUACAUAAAU ....(((.....))).((((.((((((...((((...((((.....))))....)))))))))(((((((((....)))))))))...).)))).......... ( -28.00) >DroSec_CAF1 9095 104 + 1 AAAUUCGAUUAUCGGCGGGCGCUGAAUGUACUGAAUACCAAUUUGUUUGGGAACUCGGAUUCGGAUAUACAUAUCUAUGUAUGUCACUGGGCCCAUACAUAAAU ....(((.....))).((((.((((((...((((...((((.....))))....)))))))))(((((((((....)))))))))...).)))).......... ( -28.00) >DroSim_CAF1 10118 104 + 1 AAAUUCGAUUAUCGACGGGCGCUGAAUGUGCUGAAUACCAAUUUGUUUGGGAACUCGGAUUCGGAUAUACAUAUCUAUGUAUGUCACUGGGCCCAUACAUAAAU ....(((.....))).((((.((((((...((((...((((.....))))....)))))))))(((((((((....)))))))))...).)))).......... ( -28.90) >DroEre_CAF1 11426 103 + 1 AAAUUCGAUUAUCGGCGGGCGUU-AAUGUGCUGAAUACCAAUUUGUUUGGGGACUCGGAUUCGGAUACACAUAUCAAUGUAUGUCAGUGAGUCCAUACAUAAAU ...........((((((.(....-..).))))))...((((.....))))(((((((......((((.((((....)))).))))..))))))).......... ( -25.00) >DroYak_CAF1 10564 103 + 1 AAAUUCGAUUAUCGGCGGGCGCU-AAUGUGCUGAAUACCAAUUUGUUUGGGAGCUCGGAUUCGGUUAUACAUAUCUAUGUAUGUCAGUGGGUCCAUACAUAAAU .............(((.(((((.-...))))).....((((.....))))..))).(((((((.((((((((....)))))))..).))))))).......... ( -25.50) >consensus AAAUUCGAUUAUCGGCGGGCGCUGAAUGUGCUGAAUACCAAUUUGUUUGGGAACUCGGAUUCGGAUAUACAUAUCUAUGUAUGUCACUGGGCCCAUACAUAAAU ....(((.....))).((((.(........((((...((((.....))))....)))).....(((((((((....)))))))))...).)))).......... (-24.04 = -23.88 + -0.16)


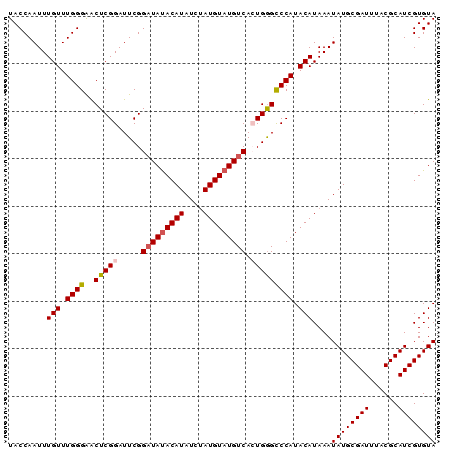
| Location | 20,870,529 – 20,870,619 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 90 |
| Reading direction | forward |
| Mean pairwise identity | 96.00 |
| Mean single sequence MFE | -25.24 |
| Consensus MFE | -22.42 |
| Energy contribution | -22.82 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.58 |
| Structure conservation index | 0.89 |
| SVM decision value | 3.46 |
| SVM RNA-class probability | 0.999244 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 20870529 90 + 27905053 UACCAAUUUGUUUGGGAACUCGGAUUCGGAUAUACAUAUCUAUGUAUGUCACUGGGCCCAUACAUAAAUAUGCGAUUUACGCAUCGUGUA ........(((.((((..(((((.....(((((((((....))))))))).))))))))).)))....((((((((......)))))))) ( -26.00) >DroSec_CAF1 9130 90 + 1 UACCAAUUUGUUUGGGAACUCGGAUUCGGAUAUACAUAUCUAUGUAUGUCACUGGGCCCAUACAUAAAUAUGCGAUUUACGCAUCGUGUA ........(((.((((..(((((.....(((((((((....))))))))).))))))))).)))....((((((((......)))))))) ( -26.00) >DroSim_CAF1 10153 90 + 1 UACCAAUUUGUUUGGGAACUCGGAUUCGGAUAUACAUAUCUAUGUAUGUCACUGGGCCCAUACAUAAAUAUGCGAUUUACGCAUCGUGUA ........(((.((((..(((((.....(((((((((....))))))))).))))))))).)))....((((((((......)))))))) ( -26.00) >DroEre_CAF1 11460 90 + 1 UACCAAUUUGUUUGGGGACUCGGAUUCGGAUACACAUAUCAAUGUAUGUCAGUGAGUCCAUACAUAAAUAUGCGAUUUACGCAUCGUGUA ..((((.....))))(((((((......((((.((((....)))).))))..))))))).(((((....(((((.....))))).))))) ( -25.20) >DroYak_CAF1 10598 90 + 1 UACCAAUUUGUUUGGGAGCUCGGAUUCGGUUAUACAUAUCUAUGUAUGUCAGUGGGUCCAUACAUAAAUAUGCGAUUUACGCAUCGUGUA ..((((.....))))......(((((((.((((((((....)))))))..).))))))).(((((....(((((.....))))).))))) ( -23.00) >consensus UACCAAUUUGUUUGGGAACUCGGAUUCGGAUAUACAUAUCUAUGUAUGUCACUGGGCCCAUACAUAAAUAUGCGAUUUACGCAUCGUGUA ........(((.((((..(((((.....(((((((((....))))))))).))))))))).)))....((((((((......)))))))) (-22.42 = -22.82 + 0.40)


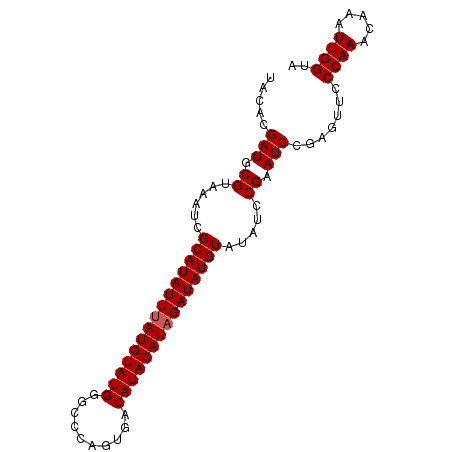
| Location | 20,870,529 – 20,870,619 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 90 |
| Reading direction | reverse |
| Mean pairwise identity | 96.00 |
| Mean single sequence MFE | -21.48 |
| Consensus MFE | -20.52 |
| Energy contribution | -20.72 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.16 |
| Structure conservation index | 0.96 |
| SVM decision value | 3.68 |
| SVM RNA-class probability | 0.999522 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 20870529 90 - 27905053 UACACGAUGCGUAAAUCGCAUAUUUAUGUAUGGGCCCAGUGACAUACAUAGAUAUGUAUAUCCGAAUCCGAGUUCCCAAACAAAUUGGUA .....(((......)))(((((((((((((((..........)))))))))))))))......((((....))))((((.....)))).. ( -21.40) >DroSec_CAF1 9130 90 - 1 UACACGAUGCGUAAAUCGCAUAUUUAUGUAUGGGCCCAGUGACAUACAUAGAUAUGUAUAUCCGAAUCCGAGUUCCCAAACAAAUUGGUA .....(((......)))(((((((((((((((..........)))))))))))))))......((((....))))((((.....)))).. ( -21.40) >DroSim_CAF1 10153 90 - 1 UACACGAUGCGUAAAUCGCAUAUUUAUGUAUGGGCCCAGUGACAUACAUAGAUAUGUAUAUCCGAAUCCGAGUUCCCAAACAAAUUGGUA .....(((......)))(((((((((((((((..........)))))))))))))))......((((....))))((((.....)))).. ( -21.40) >DroEre_CAF1 11460 90 - 1 UACACGAUGCGUAAAUCGCAUAUUUAUGUAUGGACUCACUGACAUACAUUGAUAUGUGUAUCCGAAUCCGAGUCCCCAAACAAAUUGGUA ((((..(((((.....))))).....)))).((((((...((.((((((....)))))).)).(....)))))))((((.....)))).. ( -21.50) >DroYak_CAF1 10598 90 - 1 UACACGAUGCGUAAAUCGCAUAUUUAUGUAUGGACCCACUGACAUACAUAGAUAUGUAUAACCGAAUCCGAGCUCCCAAACAAAUUGGUA .....((.((.......(((((((((((((((..........))))))))))))))).....((....)).))))((((.....)))).. ( -21.70) >consensus UACACGAUGCGUAAAUCGCAUAUUUAUGUAUGGGCCCAGUGACAUACAUAGAUAUGUAUAUCCGAAUCCGAGUUCCCAAACAAAUUGGUA .....(((.((......(((((((((((((((..........))))))))))))))).....)).))).......((((.....)))).. (-20.52 = -20.72 + 0.20)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:54:42 2006