
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 2,686,134 – 2,686,238 |
| Length | 104 |
| Max. P | 0.755642 |

| Location | 2,686,134 – 2,686,238 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.77 |
| Mean single sequence MFE | -28.24 |
| Consensus MFE | -18.29 |
| Energy contribution | -19.38 |
| Covariance contribution | 1.09 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.13 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.49 |
| SVM RNA-class probability | 0.755642 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 2686134 104 - 27905053 UCUGCGCUGGGGAAAUCAUCUUGAUGAAUUUGCCAGAUGUCUA--GAUAAAUGAAGUAAUCUUCAGUUUCUAAAAUUCACUA--------------UAGACUGAAGAGAAAGCCAGACAG ((((.(((((.(((.((((....)))).))).)).........--..............((((((((((.((........))--------------.))))))))))...)))))))... ( -26.40) >DroSec_CAF1 45538 104 - 1 UCUGCGCUGGGGAAAUCAUUUUGAUGAAUUUGCCAGAUGUCUA--GAUAAAUGAAGUAAUCUUCAGUUUCCAAAAUUCACUU--------------UAGACUGAAGAGAAAGCUAGACAG ......((((.(((.((((....)))).))).)))).((((((--(.............((((((((((.............--------------.)))))))))).....))))))). ( -28.21) >DroSim_CAF1 38691 104 - 1 UCUGCGCUGGGGAAAUCAUUUUGAUGAAUUUGCCAGAUGUCUA--GAUAAAUGAAGUAAUCUUCAGUUUCCAAAAUUCACUU--------------UAGACUGAAGAGAAAGCUAGACAG ......((((.(((.((((....)))).))).)))).((((((--(.............((((((((((.............--------------.)))))))))).....))))))). ( -28.21) >DroEre_CAF1 39522 104 - 1 UCUGCGCUGGGGAAAUCAUCUUGAUGAAUUUGCCAGAUGUCUA--GAUAAAUGAAGUAAUCUCCAGCUUCUAAACUGCACUU--------------AAGACUGAAGAGAAAGCUGGACAG ((((((((((.(((.((((....)))).))).)))).)))..)--))..............((((((((((......(....--------------..).......)).))))))))... ( -25.02) >DroYak_CAF1 46543 104 - 1 UCUGCGCUGGGGAAAUCAUCUUGAUGAAUUUGCCAGAUGUCUA--GAUAAAUGAAGUAAUCUUCAGCUUCUAAAAUUCAUCA--------------AAGACGGAAGAGAAAGCUAGACAG ......((((.(((.((((....)))).))).)))).((((((--(.....(((((....))))).(((((...........--------------.....)))))......))))))). ( -27.09) >DroAna_CAF1 32727 111 - 1 CCUCCUCUGGGGGAA--------AUCAAUCUGCCAGAUGUCCGAUGCUCGAUGAAGCAAUCUACGGCUAGAAAACUUCAUUUGGCAUGCCUCAUUCUAGGC-GAAGAGAAAGCCAGACAG .....(((((.(((.--------.....))).)))))((((.((((((.(((((((...((((....))))...))))))).)))))((((......))))-...........).)))). ( -34.50) >consensus UCUGCGCUGGGGAAAUCAUCUUGAUGAAUUUGCCAGAUGUCUA__GAUAAAUGAAGUAAUCUUCAGCUUCUAAAAUUCACUU______________UAGACUGAAGAGAAAGCUAGACAG ......((((.(((.((((....)))).))).)))).((((((............((..(((((((((..............................)))))))))....)))))))). (-18.29 = -19.38 + 1.09)
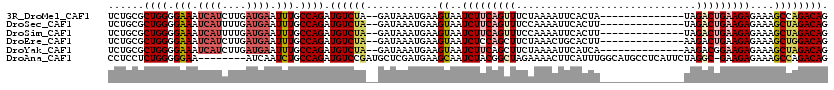
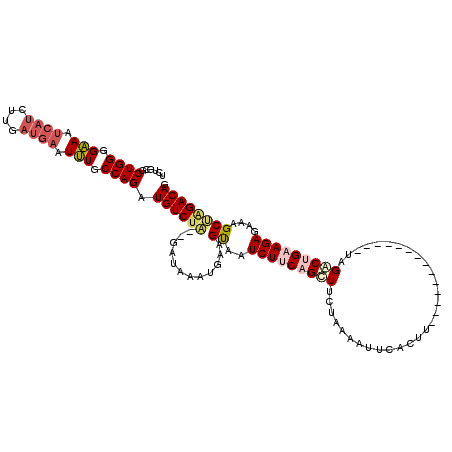
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:51:55 2006