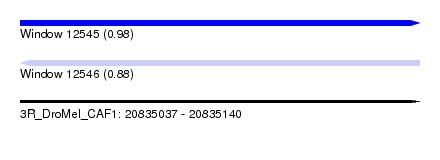
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 20,835,037 – 20,835,140 |
| Length | 103 |
| Max. P | 0.979444 |

| Location | 20,835,037 – 20,835,140 |
|---|---|
| Length | 103 |
| Sequences | 4 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 81.16 |
| Mean single sequence MFE | -33.08 |
| Consensus MFE | -20.14 |
| Energy contribution | -20.45 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.81 |
| Structure conservation index | 0.61 |
| SVM decision value | 1.84 |
| SVM RNA-class probability | 0.979444 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 20835037 103 + 27905053 UCCAAGUCGUACUCCACCCAUUCCUAUUUCUCUUUUUAUUCCUGGAUGGAGAUGG--GGAUGGAGAUGGCGGUGGAGGUGGAGGUGGAGAUGGAGGUGGGUGUGG ((((..((((.((((((((((..((((..((.((((((((((((........)))--))))))))).))..))))..)))..)))))))))))...))))..... ( -39.30) >DroSec_CAF1 50847 79 + 1 UCCAAGUAGUACUCCACCCAUUCCUAUUACUCUUUUCAUUCCUGGAUGGAGCUGG--GGAUGGA------------------------GAUGGAGGUGGGUGUGG ...........(..(((((((..(((((......((((((((..(......)..)--)))))))------------------------)))))..)))))))..) ( -29.80) >DroSim_CAF1 50298 85 + 1 UCCAAGUCGUACUCCACCCAUUCCUAUUACUCUUUUCAUUCCUGGAUGGAGCUGG--GGAUGGAGAU------------------GGAGAUGGAGGUGGGUGUGG ...........(..(((((((..(((((.((.((((((((((..(......)..)--))))))))).------------------)).)))))..)))))))..) ( -36.40) >DroYak_CAF1 44582 84 + 1 UCC-AGUCGUACUCCACCCAUUCCUAUUACUCUUCUCAUUCCUGGAUGGAGAUGGUUGGUUGGAGGU--------------------AGUUGGAGGUGGGUGUGG ...-.......(..((((((((((.(((((.(((((((((((.....))).))))......))))))--------------------))).))).)))))))..) ( -26.80) >consensus UCCAAGUCGUACUCCACCCAUUCCUAUUACUCUUUUCAUUCCUGGAUGGAGAUGG__GGAUGGAGAU____________________AGAUGGAGGUGGGUGUGG ...........(..(((((((..(((((.((((..(((((((.....)))).)))......)))).......................)))))..)))))))..) (-20.14 = -20.45 + 0.31)

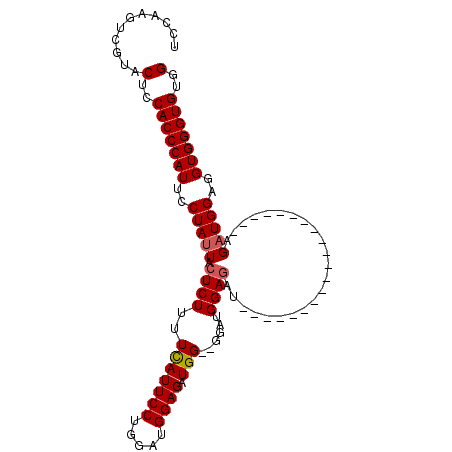

| Location | 20,835,037 – 20,835,140 |
|---|---|
| Length | 103 |
| Sequences | 4 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 81.16 |
| Mean single sequence MFE | -20.80 |
| Consensus MFE | -15.27 |
| Energy contribution | -15.53 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.92 |
| SVM RNA-class probability | 0.881549 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 20835037 103 - 27905053 CCACACCCACCUCCAUCUCCACCUCCACCUCCACCGCCAUCUCCAUCC--CCAUCUCCAUCCAGGAAUAAAAAGAGAAAUAGGAAUGGGUGGAGUACGACUUGGA ...........((((..((...((((((((...((....((((.....--.....(((.....))).......))))....))...))))))))...))..)))) ( -25.83) >DroSec_CAF1 50847 79 - 1 CCACACCCACCUCCAUC------------------------UCCAUCC--CCAGCUCCAUCCAGGAAUGAAAAGAGUAAUAGGAAUGGGUGGAGUACUACUUGGA ((.(((((((((....(------------------------((.....--.((..(((.....))).))....)))....)))..))))))(((.....))))). ( -19.00) >DroSim_CAF1 50298 85 - 1 CCACACCCACCUCCAUCUCC------------------AUCUCCAUCC--CCAGCUCCAUCCAGGAAUGAAAAGAGUAAUAGGAAUGGGUGGAGUACGACUUGGA ...........((((..((.------------------..((((((((--((.((((....((....))....))))....))...))))))))...))..)))) ( -21.60) >DroYak_CAF1 44582 84 - 1 CCACACCCACCUCCAACU--------------------ACCUCCAACCAACCAUCUCCAUCCAGGAAUGAGAAGAGUAAUAGGAAUGGGUGGAGUACGACU-GGA ((.(((((((((...(((--------------------..(((...((...............))...)))...)))...)))..)))))).((.....))-)). ( -16.76) >consensus CCACACCCACCUCCAUCU____________________AUCUCCAUCC__CCAGCUCCAUCCAGGAAUGAAAAGAGUAAUAGGAAUGGGUGGAGUACGACUUGGA ((.((((((..(((.........................................(((.....)))........(....).))).))))))(((.....))))). (-15.27 = -15.53 + 0.25)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:54:29 2006