
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 20,822,028 – 20,822,188 |
| Length | 160 |
| Max. P | 0.997869 |

| Location | 20,822,028 – 20,822,123 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 78.32 |
| Mean single sequence MFE | -32.54 |
| Consensus MFE | -17.98 |
| Energy contribution | -21.94 |
| Covariance contribution | 3.96 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.58 |
| Structure conservation index | 0.55 |
| SVM decision value | 2.06 |
| SVM RNA-class probability | 0.986943 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 20822028 95 + 27905053 AGUCCACAUAUGGCUU-------CCAGCUUCCAGCUACUGGUAUCUACUGGCUACUGGCUACUGGCUACUGACUACUGGCCAGUGGCUUCUGGUUGCAUGCC ...........(((..-------..(((.....)))....(((.(((..((((((((((((.((((....).))).))))))))))))..))).)))..))) ( -33.30) >DroSec_CAF1 37594 88 + 1 AGUCCACAUAUGGCUU-------CCAGCUUCCAGCUACUGGUAUCCACUGGCUACUGGCUACUG-------ACUACUGGCCAGUGGCUUCUGGUUGCAUGCC ...........(((..-------..(((.....)))....(((.(((..((((((((((((...-------.....))))))))))))..))).)))..))) ( -32.80) >DroSim_CAF1 37556 95 + 1 AGUCCACAUAUGGCUU-------CCAGCUUCCAGCUACUGGUAUCUACUGGCUACUGGCUACUGACUACUGACUACUGGCCAGUGGCUUCUGGUUGCAUGCC ...........(((..-------..(((.....)))....(((.(((..((((((((((((.((.........)).))))))))))))..))).)))..))) ( -29.30) >DroEre_CAF1 35403 94 + 1 AGUCCACAUAUGGCU--------CCAGCCUCCAGCUACUGGUAUCUGCUGGCUACUGGCCAGUGGCUUCUGGUUGCAUGCCGCAUGCUCCGUGCUCCAUGCU ...........(((.--------.(((((...((((((((((....(....).....))))))))))...)))))...)))(((((..........))))). ( -33.50) >DroYak_CAF1 31203 88 + 1 AGUCCACAUAUGGCUACCAGCUACCAGCUUUCAGCUACUGGUAUCCACUGGCUACUGGCUACUGGCCAGUGGC--------------UUCUGGUUGCAUGCC ...........(((((((((.....(((.....))).)))))).(((..((((((((((.....)))))))))--------------)..)))......))) ( -33.80) >consensus AGUCCACAUAUGGCUU_______CCAGCUUCCAGCUACUGGUAUCUACUGGCUACUGGCUACUGGCUACUGACUACUGGCCAGUGGCUUCUGGUUGCAUGCC ...........(((...........(((.....)))....(((.(((..((((((((((((.(((.......))).))))))))))))..))).)))..))) (-17.98 = -21.94 + 3.96)



| Location | 20,822,056 – 20,822,148 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 92 |
| Reading direction | forward |
| Mean pairwise identity | 78.37 |
| Mean single sequence MFE | -36.88 |
| Consensus MFE | -19.38 |
| Energy contribution | -23.54 |
| Covariance contribution | 4.16 |
| Combinations/Pair | 1.17 |
| Mean z-score | -3.07 |
| Structure conservation index | 0.53 |
| SVM decision value | 2.95 |
| SVM RNA-class probability | 0.997869 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
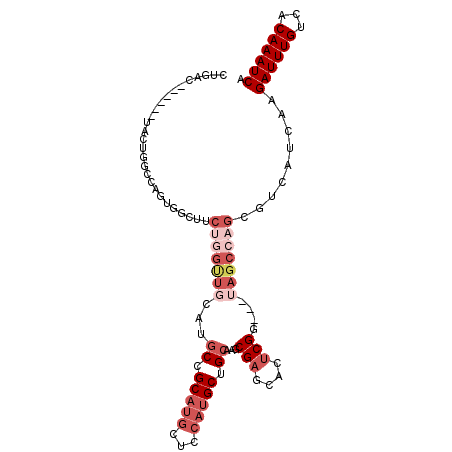
>3R_DroMel_CAF1 20822056 92 + 27905053 UACUGGUAUCUACUGGCUACUGGCUACUGGCUACUGACUACUGGCCAGUGGCUUCUGGUUGCAUGCCGCAUGCUCCACGCUGCCAACGAGCA ...(((((.(....((((((((((((.((((....).))).))))))))))))..(((..(((((...))))).))).).)))))....... ( -34.40) >DroSec_CAF1 37622 85 + 1 UACUGGUAUCCACUGGCUACUGGCUACUG-------ACUACUGGCCAGUGGCUUCUGGUUGCAUGCCGCAUGCUCCAUGCCGCCAGCGAGCA ..........(((((((((.((((....)-------.))).)))))))))(((((((((.(((((..(....)..))))).))))).)))). ( -36.70) >DroSim_CAF1 37584 92 + 1 UACUGGUAUCUACUGGCUACUGGCUACUGACUACUGACUACUGGCCAGUGGCUUCUGGUUGCAUGCCGCAUGCUCCAUGCCGCCAGCGAGCA ..........(((((((((.((((...........).))).)))))))))(((((((((.(((((..(....)..))))).))))).)))). ( -33.40) >DroEre_CAF1 35430 92 + 1 UACUGGUAUCUGCUGGCUACUGGCCAGUGGCUUCUGGUUGCAUGCCGCAUGCUCCGUGCUCCAUGCUGCAUGCUGCAUGCUGCAAGCGAGCA ..(..(...(..(((((.....)))))..)...)..)((((.(((.((((((..(((((........)))))..)))))).))).))))... ( -42.60) >DroYak_CAF1 31238 78 + 1 UACUGGUAUCCACUGGCUACUGGCUACUGGCCAGUGGC--------------UUCUGGUUGCAUGCCGCAUGCCGCAUGCUGCAAGCGAGCA .....(((.(((..((((((((((.....)))))))))--------------)..))).))).((((((.(((.(....).))).))).))) ( -37.30) >consensus UACUGGUAUCUACUGGCUACUGGCUACUGGCUACUGACUACUGGCCAGUGGCUUCUGGUUGCAUGCCGCAUGCUCCAUGCUGCCAGCGAGCA .....(((.(((..((((((((((((.(((.......))).))))))))))))..))).))).((((((..((........))..))).))) (-19.38 = -23.54 + 4.16)



| Location | 20,822,089 – 20,822,188 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 74.43 |
| Mean single sequence MFE | -32.67 |
| Consensus MFE | -12.08 |
| Energy contribution | -14.47 |
| Covariance contribution | 2.39 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.13 |
| Structure conservation index | 0.37 |
| SVM decision value | 0.29 |
| SVM RNA-class probability | 0.671554 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 20822089 99 + 27905053 CUGAC------UACUGGCCAGUGGCUUCUGGUUGCAUGCCGCAUGCUCCACGCUGCCAACGAGCACUCGGUGGUAGCCAGCGUCAUCAAGAUUUGUCACAAAUCA .((((------..((((((((......))))).(((((...))))).....(((((((.(((....))).)))))))))).))))....((((((...)))))). ( -33.90) >DroPse_CAF1 33286 86 + 1 UCCACUUCCACAUAUGGCCAG----------------CCAGCAGAUUGCAUGCUGCAGGCGAGCACACGG---UAGCCAGCAUCAUCAAGAUUUGUCACAAAUCA ..............((((..(----------------((.((((........)))).)))..((.....)---).))))..........((((((...)))))). ( -19.00) >DroSec_CAF1 37651 96 + 1 ---AC------UACUGGCCAGUGGCUUCUGGUUGCAUGCCGCAUGCUCCAUGCCGCCAGCGAGCACUCGGUGGUAGCCAGCGUCAUCAAGAUUUGUCACAAAUCA ---..------..(((((....(((...(((..(((((...))))).))).)))((((.(((....))).)))).))))).........((((((...)))))). ( -34.50) >DroSim_CAF1 37617 99 + 1 CUGAC------UACUGGCCAGUGGCUUCUGGUUGCAUGCCGCAUGCUCCAUGCCGCCAGCGAGCACUCGGUGGUAGCCAGCGUCAUCAAGAUUUGUCACAAAUCA .((((------..(((((....(((...(((..(((((...))))).))).)))((((.(((....))).)))).))))).))))....((((((...)))))). ( -38.00) >DroEre_CAF1 35463 96 + 1 CUGGU------UGCAUGCCGCAUGCUCCGUGCUCCAUGCUGCAUGCUGCAUGCUGCAAGCGAGCACUCGG---UAGCCAGCGUCAUCAAGAUUUGUCACAAAUCA (((((------(((.(((.((((((..(((((........)))))..)))))).)))..(((....))))---))))))).........((((((...)))))). ( -39.30) >DroYak_CAF1 31271 82 + 1 GUGGC--------------------UUCUGGUUGCAUGCCGCAUGCCGCAUGCUGCAAGCGAGCACUCGG---UAGCCAGCGUCAUCAAGAUUUGUCACAAAUCA (((((--------------------..((((((((.((((((.(((.(....).))).))).)))....)---))))))).)))))...((((((...)))))). ( -31.30) >consensus CUGAC______UACUGGCCAGUGGCUUCUGGUUGCAUGCCGCAUGCUCCAUGCUGCAAGCGAGCACUCGG___UAGCCAGCGUCAUCAAGAUUUGUCACAAAUCA ...........................(((((((...((.(((((...))))).))...(((....)))....))))))).........((((((...)))))). (-12.08 = -14.47 + 2.39)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:54:25 2006