
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 20,807,454 – 20,807,585 |
| Length | 131 |
| Max. P | 0.999736 |

| Location | 20,807,454 – 20,807,546 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 81.54 |
| Mean single sequence MFE | -35.28 |
| Consensus MFE | -26.19 |
| Energy contribution | -27.08 |
| Covariance contribution | 0.89 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.40 |
| SVM RNA-class probability | 0.721354 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 20807454 92 - 27905053 CU-------GAGCACUGAGAACCGAGAACGGA-UAGCAGUCGUCGCCUUCCAGCGAGGGUGAUGACUGAUGAUGCUGGCGCGACUCC-GCGUCCUCCGACC ..-------.((((.......(((....))).-...((((((((((((((....))))))))))))))....))))((((((....)-)))))........ ( -38.50) >DroPse_CAF1 17449 73 - 1 ----------------------------CGGAUUCGCCGUCGUCGCCUUCCACCGAGGGUGAUGACUGAUGAUGCUGUCGCGUCUACAGUAUUUUCCGUCU ----------------------------((((......((((((((((((....))))))))))))....((((((((.......)))))))).))))... ( -29.50) >DroSec_CAF1 23056 92 - 1 CU-------GAGCACUGAGAACCGAGAACGGA-UAGCAGUCGUCGCCUUCCAGCGAGGGUGAUGACUGAUGAUGCUGGCGCGUCUCC-GCGUCCUCCGACC ..-------.((((.......(((....))).-...((((((((((((((....))))))))))))))....))))((((((....)-)))))........ ( -38.00) >DroSim_CAF1 23007 92 - 1 CU-------GAGCACUGAGAACCGAGAACGGA-UAGCAGUCGUCGCCUUCCAGCGAGGGUGAUGACUGAUGAUGCUGGCGCGUCUCC-GCGUCCUCCGACC ..-------.((((.......(((....))).-...((((((((((((((....))))))))))))))....))))((((((....)-)))))........ ( -38.00) >DroEre_CAF1 20735 89 - 1 CU-------GAGAACUGAGAACCGAGAACGGA-UAGCAGUCGUCGCCUUCCAGCGAGGGUGAUGACUGAUGAUGCUGGCGCGACUCG-UCCUC---CGACC ..-------(((.((.(((..(((....))).-...((((((((((((((....))))))))))))))....(((....))).))))-).)))---..... ( -33.00) >DroYak_CAF1 16146 99 - 1 CUGAACACUGAGCACUGAGAACCGAGAACGGA-UAGCAGUCGUCGCCUUUCAGCGAGGGUGAUGACUGAUGAUGCUGGCGCGACUCG-UCCUCCGCCGACC ..........((((.......(((....))).-...((((((((((((((....))))))))))))))....))))((((.((....-...)))))).... ( -34.70) >consensus CU_______GAGCACUGAGAACCGAGAACGGA_UAGCAGUCGUCGCCUUCCAGCGAGGGUGAUGACUGAUGAUGCUGGCGCGACUCC_GCGUCCUCCGACC ............................((((....((((((((((((((....))))))))))))))..(((((.((.......)).))))).))))... (-26.19 = -27.08 + 0.89)

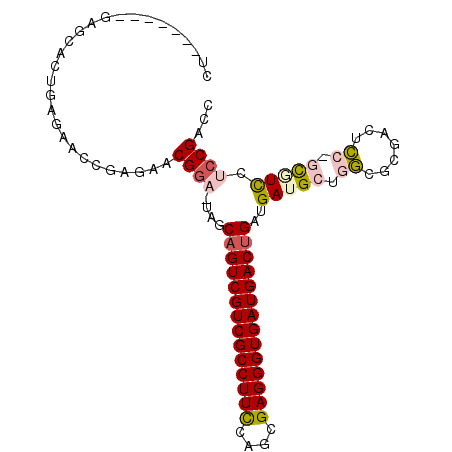

| Location | 20,807,483 – 20,807,585 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 94.75 |
| Mean single sequence MFE | -35.44 |
| Consensus MFE | -32.66 |
| Energy contribution | -32.50 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.42 |
| Structure conservation index | 0.92 |
| SVM decision value | 3.97 |
| SVM RNA-class probability | 0.999736 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 20807483 102 - 27905053 AAUCCCGUUUGCAACCCUCGAAAAACCGAGAAUCGAGAACU-------GAGCACUGAGAACCGAGAACGGAUAGCAGUCGUCGCCUUCCAGCGAGGGUGAUGACUGAUG ....((((((......((((......))))..(((.(..((-------.(....).))..))))))))))....((((((((((((((....))))))))))))))... ( -36.20) >DroSec_CAF1 23085 102 - 1 AAUCCCGUUUGCAACCCUCGAAAAACCGAGAAUCGAGAACU-------GAGCACUGAGAACCGAGAACGGAUAGCAGUCGUCGCCUUCCAGCGAGGGUGAUGACUGAUG ....((((((......((((......))))..(((.(..((-------.(....).))..))))))))))....((((((((((((((....))))))))))))))... ( -36.20) >DroSim_CAF1 23036 102 - 1 AAUCCCGUUUGCAACCCUCGAAAUAGCGAGAAUCGAGAACU-------GAGCACUGAGAACCGAGAACGGAUAGCAGUCGUCGCCUUCCAGCGAGGGUGAUGACUGAUG ....((((((......((((......))))..(((.(..((-------.(....).))..))))))))))....((((((((((((((....))))))))))))))... ( -36.00) >DroEre_CAF1 20761 101 - 1 AAUCCCGUUUGCAACCCUCGAAAAGC-GAGAAUCGAGAACU-------GAGAACUGAGAACCGAGAACGGAUAGCAGUCGUCGCCUUCCAGCGAGGGUGAUGACUGAUG ....((((((......((((.....)-)))..(((.(..((-------.(....).))..))))))))))....((((((((((((((....))))))))))))))... ( -35.80) >DroYak_CAF1 16175 109 - 1 AAUCCCGUUUGCAACCCUCGAAAAACUGAGAAUCGAGAACUGAACACUGAGCACUGAGAACCGAGAACGGAUAGCAGUCGUCGCCUUUCAGCGAGGGUGAUGACUGAUG ....(((((..(....(((((...........))))).........((.(....).))....)..)))))....((((((((((((((....))))))))))))))... ( -33.00) >consensus AAUCCCGUUUGCAACCCUCGAAAAACCGAGAAUCGAGAACU_______GAGCACUGAGAACCGAGAACGGAUAGCAGUCGUCGCCUUCCAGCGAGGGUGAUGACUGAUG ....(((((..(....(((((...........)))))..((........))...........)..)))))....((((((((((((((....))))))))))))))... (-32.66 = -32.50 + -0.16)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:54:11 2006