
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 2,682,968 – 2,683,086 |
| Length | 118 |
| Max. P | 0.994913 |

| Location | 2,682,968 – 2,683,086 |
|---|---|
| Length | 118 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.90 |
| Mean single sequence MFE | -25.95 |
| Consensus MFE | -21.43 |
| Energy contribution | -21.99 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.24 |
| SVM RNA-class probability | 0.650012 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
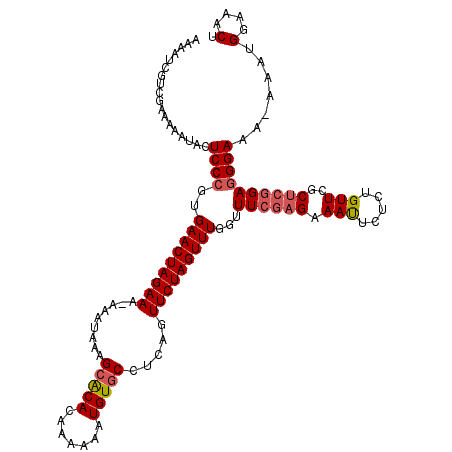
>3R_DroMel_CAF1 2682968 118 + 27905053 AAAAUCGUCGAAAAAUACUCCCGUGAACUAGAAA-AAAUAAAGCACACAAAAAAUGUGCCUCAGUUCUAGUUUAGUUUCAAGAAAUUCUCUGUUCGCUCGGAAGGAAA-AAAUGGAAACU .......................((((((((((.-.......(((((.......))))).....))))))))))(((((......(((((((......))).))))..-.....))))). ( -22.84) >DroSec_CAF1 42463 118 + 1 AAAAUCGUCAAAAAAUACUCCCGUGAACUAGAAA-AAAUAAAGCACACAAAAAAUGUGCCUCAGUUCUAGUUUGGUUUCGAGAAAUUCUCUGUUCGCUCGGAGGGAAA-AAAUGGAAACU ..................((((..(((((((((.-.......(((((.......))))).....)))))))))...((((((..(........)..))))))))))..-....(....). ( -26.82) >DroSim_CAF1 35627 118 + 1 AAAAUCGUCGAAAUAUACUCCCGUGAACUAGAAA-AAAUAAAGAACACAAAAAAUGUGCCUCAGUUCUAGUUUGGUUUCGAGAAAUUCUCUGUUCGCUCGGAGGGAAA-AAAUGGAAACU ....((.(((((((..((....))(((((((((.-......((..((((.....)))).))...))))))))).)))))))))..(((((((......)))))))...-....(....). ( -23.50) >DroEre_CAF1 36362 118 + 1 AAAAUCGUUGUAAAAUACUCCCGUGAACUAGAAA-AAAUAAAGCACACAAAAAAUGUGCCUCAGUUCUAGUUUGGUUUCGAGAAAUUCUCUGUUCGCUCGGAGGGAAA-AAAUGGGAACU ..................(((((((((((((((.-.......(((((.......))))).....)))))))))............(((((((......)))))))...-..))))))... ( -28.52) >DroYak_CAF1 43010 119 + 1 AAAAUCGUCGUAAAAUACUCCCGUGAACUAGAAAAAAAUAAAGCACACAAAAAAUGUGCCUCAGUUCUAGUUUUCUUUCGAGAAACUCUCUGUUCGCUCGGAGGGAAA-AAAUGGAAACU ..................(((((.(((((((((.........(((((.......))))).....))))))))).).((((((.(((.....)))..))))))))))..-....(....). ( -28.14) >DroAna_CAF1 29602 120 + 1 AAAAUCGUCGUAAAAUACUCCCGUGAACUAGAAAAAAAUAAAGCGCACAAAAAAUGUGCCUCAGUUCUAGUUUGGUUUCGAGAAAUUCUCUGUUCGCCCCGAGGGAAAAAAAUGGAAACU ..................((((..(((((((((........((.(((((.....)))))))...)))))))))(((..((((.....))).)...)))....)))).......(....). ( -25.90) >consensus AAAAUCGUCGAAAAAUACUCCCGUGAACUAGAAA_AAAUAAAGCACACAAAAAAUGUGCCUCAGUUCUAGUUUGGUUUCGAGAAAUUCUCUGUUCGCUCGGAGGGAAA_AAAUGGAAACU ..................((((..(((((((((.........(((((.......))))).....)))))))))...((((((.(((.....)))..)))))))))).......(....). (-21.43 = -21.99 + 0.56)

| Location | 2,682,968 – 2,683,086 |
|---|---|
| Length | 118 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.90 |
| Mean single sequence MFE | -28.31 |
| Consensus MFE | -25.94 |
| Energy contribution | -25.86 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.92 |
| SVM decision value | 2.52 |
| SVM RNA-class probability | 0.994913 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
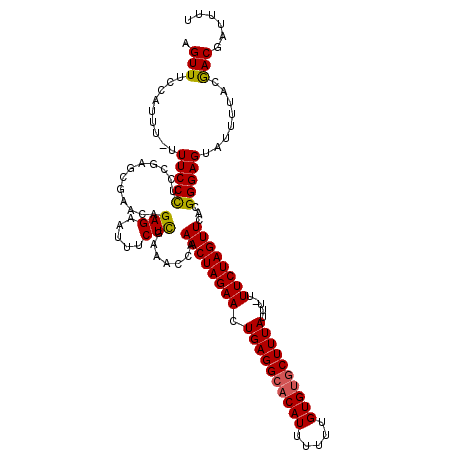
>3R_DroMel_CAF1 2682968 118 - 27905053 AGUUUCCAUUU-UUUCCUUCCGAGCGAACAGAGAAUUUCUUGAAACUAAACUAGAACUGAGGCACAUUUUUUGUGUGCUUUAUUU-UUUCUAGUUCACGGGAGUAUUUUUCGACGAUUUU ...........-....((((((........(((.....))).......((((((((.((((((((((.....))))))))))...-.))))))))..))))))................. ( -27.40) >DroSec_CAF1 42463 118 - 1 AGUUUCCAUUU-UUUCCCUCCGAGCGAACAGAGAAUUUCUCGAAACCAAACUAGAACUGAGGCACAUUUUUUGUGUGCUUUAUUU-UUUCUAGUUCACGGGAGUAUUUUUUGACGAUUUU .(((.......-.(((((..((((.((........)).))))......((((((((.((((((((((.....))))))))))...-.))))))))...)))))........)))...... ( -30.59) >DroSim_CAF1 35627 118 - 1 AGUUUCCAUUU-UUUCCCUCCGAGCGAACAGAGAAUUUCUCGAAACCAAACUAGAACUGAGGCACAUUUUUUGUGUUCUUUAUUU-UUUCUAGUUCACGGGAGUAUAUUUCGACGAUUUU .(((.......-.(((((..((((.((........)).))))......((((((((..((((((((.....)))).)))).....-.))))))))...)))))........)))...... ( -23.69) >DroEre_CAF1 36362 118 - 1 AGUUCCCAUUU-UUUCCCUCCGAGCGAACAGAGAAUUUCUCGAAACCAAACUAGAACUGAGGCACAUUUUUUGUGUGCUUUAUUU-UUUCUAGUUCACGGGAGUAUUUUACAACGAUUUU .(((.......-.(((((..((((.((........)).))))......((((((((.((((((((((.....))))))))))...-.))))))))...)))))........)))...... ( -29.49) >DroYak_CAF1 43010 119 - 1 AGUUUCCAUUU-UUUCCCUCCGAGCGAACAGAGAGUUUCUCGAAAGAAAACUAGAACUGAGGCACAUUUUUUGUGUGCUUUAUUUUUUUCUAGUUCACGGGAGUAUUUUACGACGAUUUU .(((.......-.(((((..((((.((((.....))))))))...(..((((((((.((((((((((.....)))))))))).....))))))))..))))))........)))...... ( -32.59) >DroAna_CAF1 29602 120 - 1 AGUUUCCAUUUUUUUCCCUCGGGGCGAACAGAGAAUUUCUCGAAACCAAACUAGAACUGAGGCACAUUUUUUGUGCGCUUUAUUUUUUUCUAGUUCACGGGAGUAUUUUACGACGAUUUU .(((.........(((((...((.......(((.....)))....)).((((((((..((((((((.....))))).))).......))))))))...)))))........)))...... ( -26.13) >consensus AGUUUCCAUUU_UUUCCCUCCGAGCGAACAGAGAAUUUCUCGAAACCAAACUAGAACUGAGGCACAUUUUUUGUGUGCUUUAUUU_UUUCUAGUUCACGGGAGUAUUUUACGACGAUUUU .(((.........(((((............(((.....))).......((((((((.((((((((((.....)))))))))).....))))))))...)))))........)))...... (-25.94 = -25.86 + -0.08)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:51:51 2006