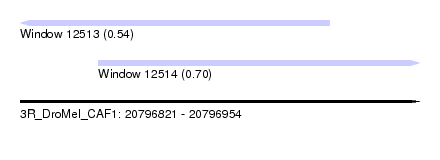
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 20,796,821 – 20,796,954 |
| Length | 133 |
| Max. P | 0.700782 |

| Location | 20,796,821 – 20,796,924 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.12 |
| Mean single sequence MFE | -21.62 |
| Consensus MFE | -17.40 |
| Energy contribution | -17.40 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.40 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.537417 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 20796821 103 - 27905053 UACAUGACGCUGCGAAUGCCAGGAACAUAAUAAAAUCAUGUAAUGGACUGCGCAUGAAAAACAAUGCAAAAA---GCGUUGACAACA------------UGGCCAUAACUGCCC--CCUC ..((((.((.((((....(((...((((.........))))..)))....)))))).....((((((.....---))))))....))------------)).............--.... ( -21.30) >DroPse_CAF1 6898 105 - 1 UACAUGACGCUGCGAAUGCCAGGAACAUAAUAAAAUCAUGUAAUGGACUGCGCAUGAAAAACAAUGCAAAAA---GCGUUGACAACA------------UGGCCAUAUUUCCCCUGCCGC ..((((.((.((((....(((...((((.........))))..)))....)))))).....((((((.....---))))))....))------------))................... ( -21.30) >DroGri_CAF1 7430 93 - 1 UACAUGACGCUGCGAAUGCCAGGAACAUAAUAAAAUCAUGUAAUGGACUGCGCAUGAAAAACAAUGCAAAAACCAGCGUUGACAAU------------CUGGU------G--------G- .......(((((.(((((((((...(((.(((......))).)))..))).))))......((((((........))))))....)------------)))))------)--------.- ( -20.40) >DroMoj_CAF1 5339 105 - 1 UACAUGACGCUGCGAAUGCCAGGAACAUAAUAAAAUCAUGUAAUGGACUGCGCAUGAAAAACAAUGCAAAAACCAGCGUUGACAACACAUGAAACGCACUGAA------G--------G- .......((.((((.(((((((...(((.(((......))).)))..))).))))......((((((........)))))).............)))).))..------.--------.- ( -22.40) >DroAna_CAF1 6332 97 - 1 UACAUGACGCUGCGAAUGCCAGGAACAUAAUAAAAUCAUGUAAUGGACUGCGCAUGAAAAACAAUGCAAAAA---GCGUUGACAACA------------UGACCAUAGGG--------UG ..((((.((.((((....(((...((((.........))))..)))....)))))).....((((((.....---))))))....))------------))(((....))--------). ( -23.00) >DroPer_CAF1 6267 105 - 1 UACAUGACGCUGCGAAUGCCAGGAACAUAAUAAAAUCAUGUAAUGGACUGCGCAUGAAAAACAAUGCAAAAA---GCGUUGACAACA------------UGGCCAUAUUUCCCCUGCCAC ..((((.((.((((....(((...((((.........))))..)))....)))))).....((((((.....---))))))....))------------))................... ( -21.30) >consensus UACAUGACGCUGCGAAUGCCAGGAACAUAAUAAAAUCAUGUAAUGGACUGCGCAUGAAAAACAAUGCAAAAA___GCGUUGACAACA____________UGGCCAUA__G________GC ..((((.(((........(((...((((.........))))..)))...))))))).....((((((........))))))....................................... (-17.40 = -17.40 + 0.00)



| Location | 20,796,847 – 20,796,954 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.76 |
| Mean single sequence MFE | -25.55 |
| Consensus MFE | -21.16 |
| Energy contribution | -21.27 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.35 |
| SVM RNA-class probability | 0.700782 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 20796847 107 + 27905053 AACGC---UUUUUGCAUUGUUUUUCAUGCGCAGUCCAUUACAUGAUUUUAUUAUGUUCCUGGCAUUCGCAGCGUCAUGUAGAAAAUGUGGACCCGACUG----CC-----CGAAUGCAA- .....---...(((((((((.......))((((((....(((((((...)))))))....((..((((((...((.....))...)))))).)))))))----).-----..)))))))- ( -25.40) >DroPse_CAF1 6926 112 + 1 AACGC---UUUUUGCAUUGUUUUUCAUGCGCAGUCCAUUACAUGAUUUUAUUAUGUUCCUGGCAUUCGCAGCGUCAUGUAGAAAAUGCAGACCCAACUG----CAACUUGCAACUGCCA- ...((---...(((((...((((((((((((.(((((..(((((((...)))))))...))).....)).))).)))).))))).(((((......)))----))...)))))..))..- ( -27.80) >DroGri_CAF1 7443 90 + 1 AACGCUGGUUUUUGCAUUGUUUUUCAUGCGCAGUCCAUUACAUGAUUUUAUUAUGUUCCUGGCAUUCGCAGCGUCAUGUAGAAAAUGCGG------------------------------ ..(((...(((((((((.(.......((((.((((((..(((((((...)))))))...))).)))))))....))))))))))..))).------------------------------ ( -24.20) >DroMoj_CAF1 5364 90 + 1 AACGCUGGUUUUUGCAUUGUUUUUCAUGCGCAGUCCAUUACAUGAUUUUAUUAUGUUCCUGGCAUUCGCAGCGUCAUGUAGAAAAUGCGG------------------------------ ..(((...(((((((((.(.......((((.((((((..(((((((...)))))))...))).)))))))....))))))))))..))).------------------------------ ( -24.20) >DroAna_CAF1 6352 112 + 1 AACGC---UUUUUGCAUUGUUUUUCAUGCGCAGUCCAUUACAUGAUUUUAUUAUGUUCCUGGCAUUCGCAGCGUCAUGUAGAAAAUGUGGACCCGACUAAAAAUU-----CAACUGCAAC .....---...((((((((..(((...(((..((((((((((((((......((((.....)))).......))))))))......)))))).)).)...)))..-----))).))))). ( -23.92) >DroPer_CAF1 6295 112 + 1 AACGC---UUUUUGCAUUGUUUUUCAUGCGCAGUCCAUUACAUGAUUUUAUUAUGUUCCUGGCAUUCGCAGCGUCAUGUAGAAAAUGCAGACCCAACUG----CAACUUGCAACUGCCA- ...((---...(((((...((((((((((((.(((((..(((((((...)))))))...))).....)).))).)))).))))).(((((......)))----))...)))))..))..- ( -27.80) >consensus AACGC___UUUUUGCAUUGUUUUUCAUGCGCAGUCCAUUACAUGAUUUUAUUAUGUUCCUGGCAUUCGCAGCGUCAUGUAGAAAAUGCGGACCC_ACUG____C______CAACUGC_A_ ..(((...(((((((((.(.......((((.((((((..(((((((...)))))))...))).)))))))....))))))))))..)))............................... (-21.16 = -21.27 + 0.11)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:53:59 2006