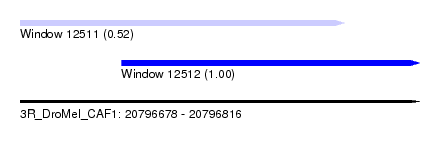
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 20,796,678 – 20,796,816 |
| Length | 138 |
| Max. P | 0.995809 |

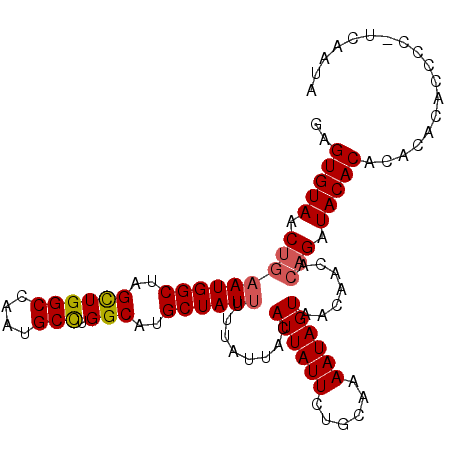
| Location | 20,796,678 – 20,796,790 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 87.27 |
| Mean single sequence MFE | -23.81 |
| Consensus MFE | -18.93 |
| Energy contribution | -20.68 |
| Covariance contribution | 1.75 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.79 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.519116 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 20796678 112 + 27905053 UACGAAAGUUAUUAUUCAAUUU-----AGUCUCAUUUUUCGAGUGUAACUGAAUGGCUAGCUGGCCAAUGCCUGGCAUGCUAUUUUUAUUACAUUAUUCUGCAAAAUAGUAACAACA .......(((((((((......-----...(((.......)))(((((..(((((((..((((((....))).)))..)))))))...)))))...........))))))))).... ( -24.90) >DroSec_CAF1 12467 112 + 1 UACGAAAGUUAUUAUUUAAUUU-----AAUCUCAUUUUUCGAGUGUAACUGAAUGGCUAGCUGGCCAAUGCCUGGCAUGCUAUUUUUAUUACAUUAUUCUGCAAAAUAGUAACAACA .......((((((((((.....-----...(((.......)))(((((..(((((((..((((((....))).)))..)))))))...)))))..........)))))))))).... ( -26.40) >DroSim_CAF1 12407 112 + 1 UACGAAAGUUAUUAUUUAAUUU-----AAUCUCAUUUUUCGAGUGUAACUGAAUGGCUAGCUGGCCAAUGCCUGGCAUGCUAUUUUUAUUACAUUAUUCUGCAAAAUAGUAACAACA .......((((((((((.....-----...(((.......)))(((((..(((((((..((((((....))).)))..)))))))...)))))..........)))))))))).... ( -26.40) >DroEre_CAF1 10082 112 + 1 UAAGAAAGAUAUUAUUUAAUUU-----GGCCUCCUUUUCCGAGUGUAACUGAAUGGCUAGCUGGCCAAUGCCUGGCAUGCUAUUUUUAUUACAUUAUUCUGCAAAAUAGUAACAACA .......(.((((((((.((((-----((.........))))))((((..(((((((..((((((....))).)))..)))))))...))))...........)))))))).).... ( -23.30) >DroYak_CAF1 4984 112 + 1 CCAUAAAGUUAUUACCUAAUUU-----AAUCUCCUUCUUCGAGUGUAACCGAAUGGCUAGCUGGCCAAUGCCUGGCAUGCUAUUUUUAUUACAUUAUUCUGCAAAAUAGUAACAACA .......(((((((........-----...(((.......)))(((((..(((((((..((((((....))).)))..)))))))...))))).............))))))).... ( -22.00) >DroAna_CAF1 6198 105 + 1 ------------UUUUUAAUUAGCUCUUGGCCCAUUUUUCAAGUGUAACUAUAUGGCUAGUUGGCCAAUGCUUGGCAUGCUAUUUUUAUUACAUUAUUCAGCAAAAUAGUAACAACA ------------........(((((..(((..((((.....))))...)))...)))))((((((((.....)))).(((((((((................))))))))).)))). ( -19.89) >consensus UACGAAAGUUAUUAUUUAAUUU_____AAUCUCAUUUUUCGAGUGUAACUGAAUGGCUAGCUGGCCAAUGCCUGGCAUGCUAUUUUUAUUACAUUAUUCUGCAAAAUAGUAACAACA .......((((((((((.............(((.......)))(((((..(((((((..((((((....))).)))..)))))))...)))))..........)))))))))).... (-18.93 = -20.68 + 1.75)

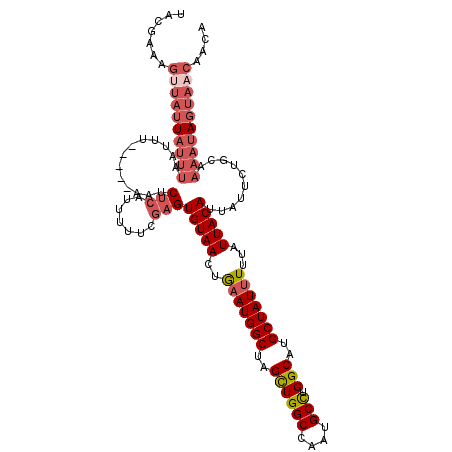
| Location | 20,796,713 – 20,796,816 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 94.90 |
| Mean single sequence MFE | -21.38 |
| Consensus MFE | -19.74 |
| Energy contribution | -19.97 |
| Covariance contribution | 0.22 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.92 |
| SVM decision value | 2.62 |
| SVM RNA-class probability | 0.995809 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 20796713 103 + 27905053 GAGUGUAACUGAAUGGCUAGCUGGCCAAUGCCUGGCAUGCUAUUUUUAUUACAUUAUUCUGCAAAAUAGUAACAACACAGAUACACACACACAACCC-UCAAUA ..(((((.(((((((((..((((((....))).)))..))))))........((((((......)))))).......))).)))))...........-...... ( -22.50) >DroSec_CAF1 12502 103 + 1 GAGUGUAACUGAAUGGCUAGCUGGCCAAUGCCUGGCAUGCUAUUUUUAUUACAUUAUUCUGCAAAAUAGUAACAACACAGAUACACACACACACCCC-UCAAUA ..(((((.(((((((((..((((((....))).)))..))))))........((((((......)))))).......))).)))))...........-...... ( -22.50) >DroSim_CAF1 12442 103 + 1 GAGUGUAACUGAAUGGCUAGCUGGCCAAUGCCUGGCAUGCUAUUUUUAUUACAUUAUUCUGCAAAAUAGUAACAACACAGAUACACACACACACCCC-UCAAUA ..(((((.(((((((((..((((((....))).)))..))))))........((((((......)))))).......))).)))))...........-...... ( -22.50) >DroEre_CAF1 10117 101 + 1 GAGUGUAACUGAAUGGCUAGCUGGCCAAUGCCUGGCAUGCUAUUUUUAUUACAUUAUUCUGCAAAAUAGUAACAACACAGAUACACACACAC--CCC-UCAAUA ..(((((.(((((((((..((((((....))).)))..))))))........((((((......)))))).......))).)))))......--...-...... ( -22.50) >DroYak_CAF1 5019 102 + 1 GAGUGUAACCGAAUGGCUAGCUGGCCAAUGCCUGGCAUGCUAUUUUUAUUACAUUAUUCUGCAAAAUAGUAACAACACAGAUACACACACAC--CCCGUCAAUA ..(((((...(((((((..((((((....))).)))..)))))))............((((................)))))))))......--.......... ( -19.99) >DroAna_CAF1 6226 103 + 1 AAGUGUAACUAUAUGGCUAGUUGGCCAAUGCUUGGCAUGCUAUUUUUAUUACAUUAUUCAGCAAAAUAGUAACAACACAGAUACACACACACACCUC-UGCACA ..(((((.((...(((((....))))).((.(((...(((((((((................))))))))).))))).)).)))))...........-...... ( -18.29) >consensus GAGUGUAACUGAAUGGCUAGCUGGCCAAUGCCUGGCAUGCUAUUUUUAUUACAUUAUUCUGCAAAAUAGUAACAACACAGAUACACACACACACCCC_UCAAUA ..(((((.(((((((((..((((((....))).)))..))))))........((((((......)))))).......))).))))).................. (-19.74 = -19.97 + 0.22)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:53:57 2006