
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 20,788,724 – 20,789,032 |
| Length | 308 |
| Max. P | 0.988546 |

| Location | 20,788,724 – 20,788,844 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.06 |
| Mean single sequence MFE | -34.95 |
| Consensus MFE | -32.28 |
| Energy contribution | -32.15 |
| Covariance contribution | -0.13 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.41 |
| Structure conservation index | 0.92 |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.538770 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 20788724 120 - 27905053 CGAGGCGGUCUAGAUGUGAAAGGCUUUUGAAUUUGGCUAAAAUAUGUAUGUAUGUUUGGACGCUUCAUUCGGGUGUUGAAGAAGCUUUCACGCCGCCCAAAAAACAAAGACCCCAAAUCG ...((.(((((...(((....(((...(((((..(((..(((((((....)))))))....)))..)))))(((((.((((....))))))))))))......))).)))))))...... ( -32.00) >DroSec_CAF1 4534 116 - 1 CGAGGCGGUCUAGAUGUGAAAGGCUUUUGAAUUUGGCUAAAAUAUGU----AUGUUUGGCCGCUCCAUUCGGGCGAUGAAGAAGCUUUCACGCCGCCCAAAAAACAAAGACCCCAAAUCG .(.((((((......(((((((.(((((..(((.(((((((......----...)))))))((((.....)))))))..)))))))))))))))))))...................... ( -35.10) >DroSim_CAF1 4489 116 - 1 CGAGGCGGUCUAGAUGUGAAAGGCUUUUGAAUUUGGCUAAAAUAUGU----AUGUUUGGCCGCUCCAUUCGGGCGGUGAAGAAGCUUUCACGCCGCCCAAAAAACAAAGACCCCAAAUCG ...((.(((((.((((.((..((((...((((...((........))----..))))))))..)))))).((((((((..((.....)).)))))))).........)))))))...... ( -37.60) >DroEre_CAF1 1148 114 - 1 CGAGGCGGUCUAGAUGUGAAAGGCUUUUGAAUUUGGCUAAAAUAUGU----AUGUUUGGCCGCUCCAUUCGGGCGAUGAAGAAGCUUUCACGCCGCCCAA--AACAAAGGCCCCAAAUCG .(.((((((......(((((((.(((((..(((.(((((((......----...)))))))((((.....)))))))..)))))))))))))))))))..--.................. ( -35.10) >consensus CGAGGCGGUCUAGAUGUGAAAGGCUUUUGAAUUUGGCUAAAAUAUGU____AUGUUUGGCCGCUCCAUUCGGGCGAUGAAGAAGCUUUCACGCCGCCCAAAAAACAAAGACCCCAAAUCG ...((.(((((...(((....(((...(((((.((((((((.(((......)))))))))))....)))))((((.(((((....))))))))))))......))).)))))))...... (-32.28 = -32.15 + -0.13)



| Location | 20,788,804 – 20,788,924 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.50 |
| Mean single sequence MFE | -37.55 |
| Consensus MFE | -34.15 |
| Energy contribution | -34.65 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.10 |
| Structure conservation index | 0.91 |
| SVM decision value | 0.68 |
| SVM RNA-class probability | 0.819869 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 20788804 120 - 27905053 UAAGCCCAUAAGCCUUUUAGAUUUGCAAUUGUCCGCGAGCGUUUGAGCCCAAUUCUCCAGAGAUUCUGACAAAACAUCAGCGAGGCGGUCUAGAUGUGAAAGGCUUUUGAAUUUGGCUAA ..((((((.((((((((((.(((((..((((((((((((...(((....)))..)))(((.....)))...........))).)))))).))))).)))))))))).)).....)))).. ( -36.70) >DroSec_CAF1 4610 120 - 1 UAAGCCCAUAAGCCUUUUAGCUUUGCAAUUGUCCGCGAGCGCUUGCGCCCAAUUCUCCAGAGAUUCUGACAAAACAUCAGCGAGGCGGUCUAGAUGUGAAAGGCUUUUGAAUUUGGCUAA ..((((((.((((((((((((((((((((((..(((((....)))))..)))))...(((.....)))...........))))))).((....)).)))))))))).)).....)))).. ( -37.80) >DroSim_CAF1 4565 120 - 1 UAAGCCCAUAAGCCUUUUAGCUUUGCAAUUGUCCGCGAGCGCUUGCGCCCAAUUCUCCAGAGAUUCUGACAAAACAUCAGCGAGGCGGUCUAGAUGUGAAAGGCUUUUGAAUUUGGCUAA ..((((((.((((((((((((((((((((((..(((((....)))))..)))))...(((.....)))...........))))))).((....)).)))))))))).)).....)))).. ( -37.80) >DroEre_CAF1 1222 120 - 1 UAAGCCCAUAAGCCUUUUAGCUUUGCAAUUGUCCGCGAGCGCUUGCGCCCAAUUCUCCAGAGAUUCUGACAAAACACUCGCGAGGCGGUCUAGAUGUGAAAGGCUUUUGAAUUUGGCUAA ..((((((.((((((((((((((((((((((..(((((....)))))..)))))...(((.....)))...........))))))).((....)).)))))))))).)).....)))).. ( -37.90) >consensus UAAGCCCAUAAGCCUUUUAGCUUUGCAAUUGUCCGCGAGCGCUUGCGCCCAAUUCUCCAGAGAUUCUGACAAAACAUCAGCGAGGCGGUCUAGAUGUGAAAGGCUUUUGAAUUUGGCUAA ..((((((.((((((((((((((((((((((..(((((....)))))..)))))...(((.....)))...........))))))).((....)).)))))))))).)).....)))).. (-34.15 = -34.65 + 0.50)



| Location | 20,788,884 – 20,788,992 |
|---|---|
| Length | 108 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.52 |
| Mean single sequence MFE | -43.23 |
| Consensus MFE | -38.77 |
| Energy contribution | -39.27 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.56 |
| Structure conservation index | 0.90 |
| SVM decision value | 2.12 |
| SVM RNA-class probability | 0.988546 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
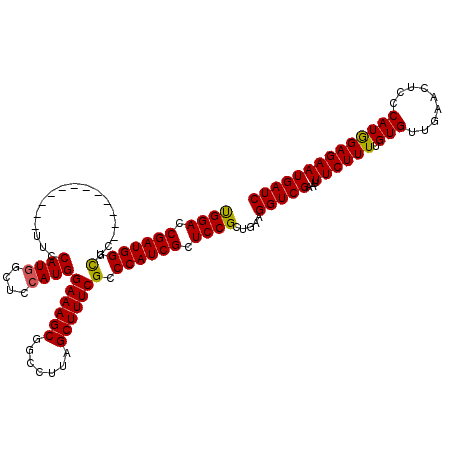
>3R_DroMel_CAF1 20788884 108 + 27905053 GCUCGCGGACAAUUGCAAAUCUAAAAGGCUUAUGGGCUUAUGGACCGAUGGUCGC------------UUCCAUGGCUCCAUGGAAAGCGGCCUUAGCUUUCGCCCAUCGCUCCGCUGAAG ....((((((....((...........))..((((((....((.(..(.((((((------------(((((((....))))).)))))))))..)))...)))))).).)))))..... ( -40.00) >DroSec_CAF1 4690 108 + 1 GCUCGCGGACAAUUGCAAAGCUAAAAGGCUUAUGGGCUUAUGGAUCGAUGGUCGC------------UUCCAUGGCUCCAUGGAAAGCGGCCUUAGCUUUCGCCCAUCGCUCCGCUGAAG ....(((((.....((.(((((....)))))((((((....((......((((((------------(((((((....))))).))))))))....))...)))))).)))))))..... ( -44.50) >DroSim_CAF1 4645 108 + 1 GCUCGCGGACAAUUGCAAAGCUAAAAGGCUUAUGGGCUUAUGGAUCGAUGGUCGC------------UUCCAUGGCUCCAUGGAAAGCGGCCUUAGCUUUCGCCCAUCGCUCCGCUGAAG ....(((((.....((.(((((....)))))((((((....((......((((((------------(((((((....))))).))))))))....))...)))))).)))))))..... ( -44.50) >DroEre_CAF1 1302 120 + 1 GCUCGCGGACAAUUGCAAAGCUAAAAGGCUUAUGGGCUUACGGACCGAUGGUUCCAUAGCUCCAUGGCUCCAUAGCUCCAUGGAAAGCGGCCUUAGCUUUCGCCCAUCGCUCCGCUGAAG ....(((((.....((.(((((....)))))((((((....(((.(.((((..((((......))))..)))).).)))...((((((.......)))))))))))).)))))))..... ( -43.90) >consensus GCUCGCGGACAAUUGCAAAGCUAAAAGGCUUAUGGGCUUAUGGACCGAUGGUCGC____________UUCCAUGGCUCCAUGGAAAGCGGCCUUAGCUUUCGCCCAUCGCUCCGCUGAAG ....(((((.....((.(((((....)))))((((((.((((((.(.((((..................)))).).))))))((((((.......)))))))))))).)))))))..... (-38.77 = -39.27 + 0.50)


| Location | 20,788,884 – 20,788,992 |
|---|---|
| Length | 108 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.52 |
| Mean single sequence MFE | -43.33 |
| Consensus MFE | -35.98 |
| Energy contribution | -36.05 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.58 |
| Structure conservation index | 0.83 |
| SVM decision value | 1.54 |
| SVM RNA-class probability | 0.962733 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 20788884 108 - 27905053 CUUCAGCGGAGCGAUGGGCGAAAGCUAAGGCCGCUUUCCAUGGAGCCAUGGAA------------GCGACCAUCGGUCCAUAAGCCCAUAAGCCUUUUAGAUUUGCAAUUGUCCGCGAGC .....(((((.((((..(((((....((((((((((.(((((....)))))))------------)))......(((......))).....))))).....))))).))))))))).... ( -40.30) >DroSec_CAF1 4690 108 - 1 CUUCAGCGGAGCGAUGGGCGAAAGCUAAGGCCGCUUUCCAUGGAGCCAUGGAA------------GCGACCAUCGAUCCAUAAGCCCAUAAGCCUUUUAGCUUUGCAAUUGUCCGCGAGC .....(((((.((((((((....))......(((((.(((((....)))))))------------))).))))))........((....((((......)))).)).....))))).... ( -42.00) >DroSim_CAF1 4645 108 - 1 CUUCAGCGGAGCGAUGGGCGAAAGCUAAGGCCGCUUUCCAUGGAGCCAUGGAA------------GCGACCAUCGAUCCAUAAGCCCAUAAGCCUUUUAGCUUUGCAAUUGUCCGCGAGC .....(((((.((((((((....))......(((((.(((((....)))))))------------))).))))))........((....((((......)))).)).....))))).... ( -42.00) >DroEre_CAF1 1302 120 - 1 CUUCAGCGGAGCGAUGGGCGAAAGCUAAGGCCGCUUUCCAUGGAGCUAUGGAGCCAUGGAGCUAUGGAACCAUCGGUCCGUAAGCCCAUAAGCCUUUUAGCUUUGCAAUUGUCCGCGAGC .....(((((.((((..((.(((((((((((.....(((((((..(....)..)))))))((((((((.((...))))))).)))......))))..))))))))).))))))))).... ( -49.00) >consensus CUUCAGCGGAGCGAUGGGCGAAAGCUAAGGCCGCUUUCCAUGGAGCCAUGGAA____________GCGACCAUCGAUCCAUAAGCCCAUAAGCCUUUUAGCUUUGCAAUUGUCCGCGAGC .....((((((((((((((((((((.......)))))).(((((.(.((((..................)))).).)))))..))))))((((......))))))).....))))).... (-35.98 = -36.05 + 0.06)


| Location | 20,788,924 – 20,789,032 |
|---|---|
| Length | 108 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.50 |
| Mean single sequence MFE | -37.70 |
| Consensus MFE | -29.16 |
| Energy contribution | -28.85 |
| Covariance contribution | -0.31 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.45 |
| SVM RNA-class probability | 0.740088 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 20788924 108 + 27905053 UGGACCGAUGGUCGC------------UUCCAUGGCUCCAUGGAAAGCGGCCUUAGCUUUCGCCCAUCGCUCCGCUGAAGGUCGAAUUUCUUUUGUGUUGAACUCCCAUAGAGAAUGAUC ..((((...((((((------------(((((((....))))).))))))))(((((....((.....))...))))).))))....((((((.(((.........)))))))))..... ( -35.80) >DroSec_CAF1 4730 108 + 1 UGGAUCGAUGGUCGC------------UUCCAUGGCUCCAUGGAAAGCGGCCUUAGCUUUCGCCCAUCGCUCCGCUGAAGGUCGAAUUUCUUUUGUGUUGAACCUCCAUGGAGAAUGAUC ..(((((((((((((------------(((((((....))))).))))))))...((....))..))).(((((.((.(((((((....(....)..))).)))).)))))))...)))) ( -38.90) >DroSim_CAF1 4685 108 + 1 UGGAUCGAUGGUCGC------------UUCCAUGGCUCCAUGGAAAGCGGCCUUAGCUUUCGCCCAUCGCUCCGCUGAAGGUCGAAUUUCUUUUGUGUUGAACUUCCAUGGAGAAUGAUC ..(((((((((((((------------(((((((....))))).))))))))...((....))..))).(((((..((((.((((....(....)..)))).))))..)))))...)))) ( -39.00) >DroEre_CAF1 1342 120 + 1 CGGACCGAUGGUUCCAUAGCUCCAUGGCUCCAUAGCUCCAUGGAAAGCGGCCUUAGCUUUCGCCCAUCGCUCCGCUGAAGGUCGAAUUUCUUUUGUGUUGAACUCCCAUGGAGAAUGAUC .(((((...)))))(((..((((((((.....((((.(...(((((.((((((((((....((.....))...)))).))))))..)))))...).)))).....)))))))).)))... ( -37.10) >consensus UGGACCGAUGGUCGC____________UUCCAUGGCUCCAUGGAAAGCGGCCUUAGCUUUCGCCCAUCGCUCCGCUGAAGGUCGAAUUUCUUUUGUGUUGAACUCCCAUGGAGAAUGAUC ((((.((((((.(.................((((....))))((((((.......))))))).)))))).)))).....(((((...((((((.(((.........)))))))))))))) (-29.16 = -28.85 + -0.31)


| Location | 20,788,924 – 20,789,032 |
|---|---|
| Length | 108 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.50 |
| Mean single sequence MFE | -41.66 |
| Consensus MFE | -34.22 |
| Energy contribution | -33.34 |
| Covariance contribution | -0.88 |
| Combinations/Pair | 1.13 |
| Mean z-score | -3.29 |
| Structure conservation index | 0.82 |
| SVM decision value | 1.94 |
| SVM RNA-class probability | 0.983451 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 20788924 108 - 27905053 GAUCAUUCUCUAUGGGAGUUCAACACAAAAGAAAUUCGACCUUCAGCGGAGCGAUGGGCGAAAGCUAAGGCCGCUUUCCAUGGAGCCAUGGAA------------GCGACCAUCGGUCCA ((..(((((.....))))))).........(((........)))...(((.((((((((....))......(((((.(((((....)))))))------------))).)))))).))). ( -39.10) >DroSec_CAF1 4730 108 - 1 GAUCAUUCUCCAUGGAGGUUCAACACAAAAGAAAUUCGACCUUCAGCGGAGCGAUGGGCGAAAGCUAAGGCCGCUUUCCAUGGAGCCAUGGAA------------GCGACCAUCGAUCCA ((((...((((.((((((((.................))))))))..)))).(((((((....))......(((((.(((((....)))))))------------))).))))))))).. ( -44.83) >DroSim_CAF1 4685 108 - 1 GAUCAUUCUCCAUGGAAGUUCAACACAAAAGAAAUUCGACCUUCAGCGGAGCGAUGGGCGAAAGCUAAGGCCGCUUUCCAUGGAGCCAUGGAA------------GCGACCAUCGAUCCA ((((...((((...((((.((................)).))))...)))).(((((((....))......(((((.(((((....)))))))------------))).))))))))).. ( -39.69) >DroEre_CAF1 1342 120 - 1 GAUCAUUCUCCAUGGGAGUUCAACACAAAAGAAAUUCGACCUUCAGCGGAGCGAUGGGCGAAAGCUAAGGCCGCUUUCCAUGGAGCUAUGGAGCCAUGGAGCUAUGGAACCAUCGGUCCG ((..(((((.....))))))).........(((........)))..((((.((((((((....))..........((((((((..(((((....)))))..)))))))))))))).)))) ( -43.00) >consensus GAUCAUUCUCCAUGGAAGUUCAACACAAAAGAAAUUCGACCUUCAGCGGAGCGAUGGGCGAAAGCUAAGGCCGCUUUCCAUGGAGCCAUGGAA____________GCGACCAUCGAUCCA ((((...((((((((((.............(((........)))(((((..(....(((....)))..).)))))))))))))))..((((..................)))).)))).. (-34.22 = -33.34 + -0.88)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:53:52 2006