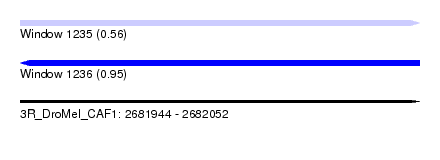
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 2,681,944 – 2,682,052 |
| Length | 108 |
| Max. P | 0.946500 |

| Location | 2,681,944 – 2,682,052 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 85.49 |
| Mean single sequence MFE | -20.68 |
| Consensus MFE | -14.27 |
| Energy contribution | -14.97 |
| Covariance contribution | 0.70 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.84 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.559887 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
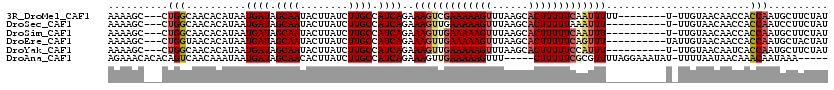
>3R_DroMel_CAF1 2681944 108 + 27905053 AAAAGC---CUGGCAACACAUAAUGAUAGCAAUACUUAUCUUGCCAUCAGAAAGUCGAAAAAGUUUAAGCACUUUUUCAAUUUUU--------U-UUGUAACAACCACCAAUGCUUCUAU ..((((---.(((..........((((.((((........)))).))))((((((.((((((((......)))))))).))))))--------.-............)))..)))).... ( -20.10) >DroSec_CAF1 41520 106 + 1 AAAAGC---CUGGCAACACAUAAUGAUAGCAAUACUUAUCUUGCCAUCAGAAAGUUGAAAAAGUUUAAGCACUUUUUAAAUUU----------U-UUGUAACAACCACCAAUCCUUCUAU ......---.((((((........(((((......))))))))))).(((((((((.(((((((......))))))).)))))----------)-)))...................... ( -17.30) >DroSim_CAF1 34681 106 + 1 AAAAGC---CUGGCAACACAUAAUGAUAGCAAUACUUAUCUUGCCAUCAGAAAGUUGAAAAAGUUUAAGCACUUUUUCAAUUU----------U-UUGUAACAACCACCAAUGCUUCUAU ..((((---.((((((........(((((......))))))))))).(((((((((((((((((......)))))))))))))----------)-)))..............)))).... ( -26.00) >DroEre_CAF1 35384 107 + 1 AAAAGC---CUGGUAACACAUAAUGAUAGCAAUACUUAUCUUGCCAUCAGAAAGUUGAAAAAGUUUAAGCACUUUUUCAGUUU----------UAUUGUAACAACCACCAAUGCUACUAU ...(((---.((((.........((((.((((........)))).)))).(((..(((((((((......)))))))))..))----------)............))))..)))..... ( -22.30) >DroYak_CAF1 41985 106 + 1 AAAAGC---CUGGCAACACAUAAUGAUAGCAAUACUUAUCUUGCCAUCAGAAAGUUGAAAAAGUUUAAGCACUUUUUCCAUUU----------U-UUGUAACAAUCACCAAUGCUUCUAU ..((((---.((((((........(((((......))))))))))).((((((((.((((((((......)))))))).))))----------)-)))..............)))).... ( -21.90) >DroAna_CAF1 29038 109 + 1 AGAAACACACAGUCAACAAAUAAUGAUAGCAACACUUAUCUUGCCAUCAGAAAGUUGAAAAAGUUU-----CUUUUUCGCGUUUUAGGAAAUAU-UUUUAAUAACAAACAAUAAA----- ((((((......(((((......((((.((((........)))).))))....)))))....))))-----))(((((........)))))...-....................----- ( -16.50) >consensus AAAAGC___CUGGCAACACAUAAUGAUAGCAAUACUUAUCUUGCCAUCAGAAAGUUGAAAAAGUUUAAGCACUUUUUCAAUUU__________U_UUGUAACAACCACCAAUGCUUCUAU ..........(((..........((((.((((........)))).))))..(((((((((((((......)))))))))))))........................))).......... (-14.27 = -14.97 + 0.70)
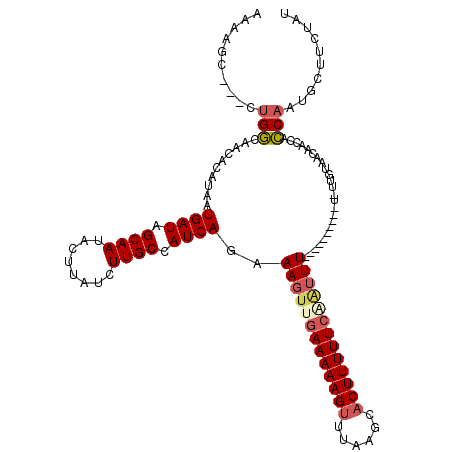
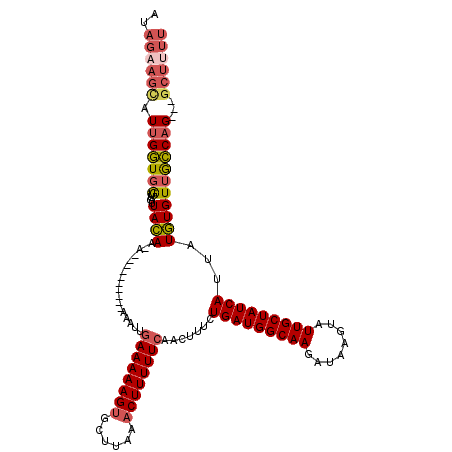
| Location | 2,681,944 – 2,682,052 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 85.49 |
| Mean single sequence MFE | -28.83 |
| Consensus MFE | -23.42 |
| Energy contribution | -24.32 |
| Covariance contribution | 0.89 |
| Combinations/Pair | 1.15 |
| Mean z-score | -3.24 |
| Structure conservation index | 0.81 |
| SVM decision value | 1.36 |
| SVM RNA-class probability | 0.946500 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 2681944 108 - 27905053 AUAGAAGCAUUGGUGGUUGUUACAA-A--------AAAAAUUGAAAAAGUGCUUAAACUUUUUCGACUUUCUGAUGGCAAGAUAAGUAUUGCUAUCAUUAUGUGUUGCCAG---GCUUUU ..((((((.((((..(....((((.-.--------.....((((((((((......)))))))))).....(((((((((........)))))))))...)))))..))))---)))))) ( -33.00) >DroSec_CAF1 41520 106 - 1 AUAGAAGGAUUGGUGGUUGUUACAA-A----------AAAUUUAAAAAGUGCUUAAACUUUUUCAACUUUCUGAUGGCAAGAUAAGUAUUGCUAUCAUUAUGUGUUGCCAG---GCUUUU ...(((((.((((..(....((((.-.----------......(((((((......)))))))........(((((((((........)))))))))...)))))..))))---.))))) ( -24.70) >DroSim_CAF1 34681 106 - 1 AUAGAAGCAUUGGUGGUUGUUACAA-A----------AAAUUGAAAAAGUGCUUAAACUUUUUCAACUUUCUGAUGGCAAGAUAAGUAUUGCUAUCAUUAUGUGUUGCCAG---GCUUUU ..((((((.((((..(....((((.-.----------...((((((((((......)))))))))).....(((((((((........)))))))))...)))))..))))---)))))) ( -33.30) >DroEre_CAF1 35384 107 - 1 AUAGUAGCAUUGGUGGUUGUUACAAUA----------AAACUGAAAAAGUGCUUAAACUUUUUCAACUUUCUGAUGGCAAGAUAAGUAUUGCUAUCAUUAUGUGUUACCAG---GCUUUU .....(((.(((((((....((((...----------....(((((((((......)))))))))......(((((((((........)))))))))...)))))))))))---)))... ( -29.60) >DroYak_CAF1 41985 106 - 1 AUAGAAGCAUUGGUGAUUGUUACAA-A----------AAAUGGAAAAAGUGCUUAAACUUUUUCAACUUUCUGAUGGCAAGAUAAGUAUUGCUAUCAUUAUGUGUUGCCAG---GCUUUU ..((((((.((((..((....(((.-.----------.....((((((((......)))))))).......(((((((((........)))))))))...)))))..))))---)))))) ( -32.10) >DroAna_CAF1 29038 109 - 1 -----UUUAUUGUUUGUUAUUAAAA-AUAUUUCCUAAAACGCGAAAAAG-----AAACUUUUUCAACUUUCUGAUGGCAAGAUAAGUGUUGCUAUCAUUAUUUGUUGACUGUGUGUUUCU -----.....(((((........))-))).......(((((((((((((-----...))))))).......(((((((((.(....).)))))))))...............)))))).. ( -20.30) >consensus AUAGAAGCAUUGGUGGUUGUUACAA_A__________AAAUUGAAAAAGUGCUUAAACUUUUUCAACUUUCUGAUGGCAAGAUAAGUAUUGCUAUCAUUAUGUGUUGCCAG___GCUUUU ..((((((.(((((((....((((..................((((((((......)))))))).......(((((((((........)))))))))...)))))))))))...)))))) (-23.42 = -24.32 + 0.89)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:51:49 2006