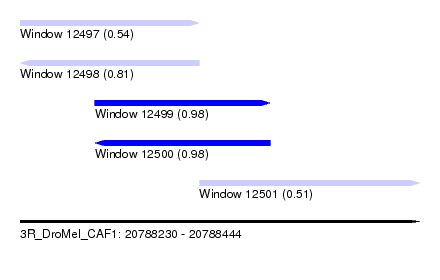
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 20,788,230 – 20,788,444 |
| Length | 214 |
| Max. P | 0.980176 |

| Location | 20,788,230 – 20,788,326 |
|---|---|
| Length | 96 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 85.65 |
| Mean single sequence MFE | -22.62 |
| Consensus MFE | -13.89 |
| Energy contribution | -14.95 |
| Covariance contribution | 1.06 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.541135 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
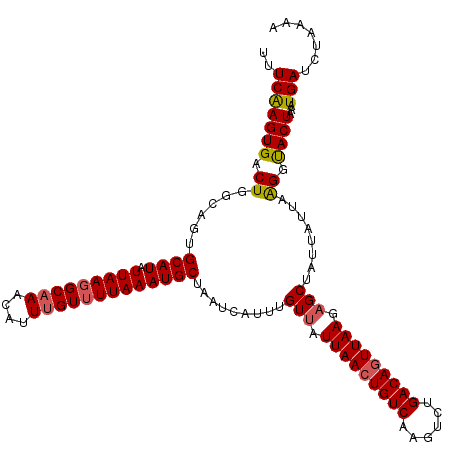
>3R_DroMel_CAF1 20788230 96 + 27905053 AAUUGGCCACCUAAACAAGCCGAAAACAUAUUCACAACAUUGCCCACAUUGAUAAGGGUCACAAAUUAAAUGACCCGUCGUGAGCACUGAGAGAAA------------------------ ..(((((...........)))))..............((.(((.(((...(((..((((((.........)))))))))))).))).)).......------------------------ ( -20.20) >DroSec_CAF1 4042 96 + 1 AAUUGGCCACCUAAACAAGCCGAAAACAUAUUCACAACAUUGCCCACAUUGAUAAGGGUCACAAAUUAAAUGACCCGUCGAGAGCACUGAGAGAAA------------------------ ..(((((...........)))))..............((.(((.....(((((..((((((.........)))))))))))..))).)).......------------------------ ( -18.80) >DroSim_CAF1 3997 96 + 1 AAUUGGCCACCUAAACAAGCCGAAAACAUAUUCACAACAUUGCCCACAUUGAUAAGGGUCACAAAUUAAAUGACCCGACGUGAGCACUGAGAGAAA------------------------ ..(((((...........)))))..............((.(((.(((........((((((.........))))))...))).))).)).......------------------------ ( -18.90) >DroEre_CAF1 661 120 + 1 AAUUGGGCACCUAAACAAGCCCAAAACAUAUUCACAACAUUGCCCACAUUGAUAAGGGUCACAAAUUAAGUGAGCUGUCGUGAGCACUGAGAGAAAAUGGGGCAGCAAACUGCGUGUUUG ...(((((..........)))))((((((...........(((((.((((.....((.((((.......)))).))(((....).))........)))))))))((.....)))))))). ( -32.60) >consensus AAUUGGCCACCUAAACAAGCCGAAAACAUAUUCACAACAUUGCCCACAUUGAUAAGGGUCACAAAUUAAAUGACCCGUCGUGAGCACUGAGAGAAA________________________ ..(((((...........)))))..............((.(((.(((...(((..((((((.........)))))))))))).))).))............................... (-13.89 = -14.95 + 1.06)

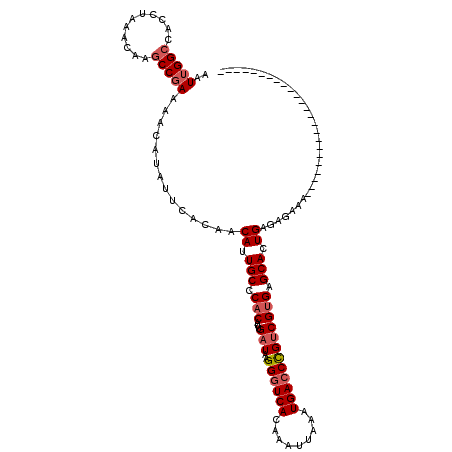
| Location | 20,788,230 – 20,788,326 |
|---|---|
| Length | 96 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 85.65 |
| Mean single sequence MFE | -27.75 |
| Consensus MFE | -21.84 |
| Energy contribution | -22.65 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.65 |
| SVM RNA-class probability | 0.813063 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 20788230 96 - 27905053 ------------------------UUUCUCUCAGUGCUCACGACGGGUCAUUUAAUUUGUGACCCUUAUCAAUGUGGGCAAUGUUGUGAAUAUGUUUUCGGCUUGUUUAGGUGGCCAAUU ------------------------.(((.(.((.((((((((..(((((((.......))))))).......)))))))).))..).))).........((((.........)))).... ( -27.10) >DroSec_CAF1 4042 96 - 1 ------------------------UUUCUCUCAGUGCUCUCGACGGGUCAUUUAAUUUGUGACCCUUAUCAAUGUGGGCAAUGUUGUGAAUAUGUUUUCGGCUUGUUUAGGUGGCCAAUU ------------------------.(((.(.((.(((((.((..(((((((.......))))))).......)).))))).))..).))).........((((.........)))).... ( -23.10) >DroSim_CAF1 3997 96 - 1 ------------------------UUUCUCUCAGUGCUCACGUCGGGUCAUUUAAUUUGUGACCCUUAUCAAUGUGGGCAAUGUUGUGAAUAUGUUUUCGGCUUGUUUAGGUGGCCAAUU ------------------------.(((.(.((.(((((((((.(((((((.......)))))))......))))))))).))..).))).........((((.........)))).... ( -28.70) >DroEre_CAF1 661 120 - 1 CAAACACGCAGUUUGCUGCCCCAUUUUCUCUCAGUGCUCACGACAGCUCACUUAAUUUGUGACCCUUAUCAAUGUGGGCAAUGUUGUGAAUAUGUUUUGGGCUUGUUUAGGUGCCCAAUU .((((..((((....))))..............(((.((((((((((((((........(((......)))..))))))..)))))))).)))))))(((((..........)))))... ( -32.10) >consensus ________________________UUUCUCUCAGUGCUCACGACGGGUCAUUUAAUUUGUGACCCUUAUCAAUGUGGGCAAUGUUGUGAAUAUGUUUUCGGCUUGUUUAGGUGGCCAAUU .........................(((.(.((.((((((((..(((((((.......))))))).......)))))))).))..).))).........((((.........)))).... (-21.84 = -22.65 + 0.81)



| Location | 20,788,270 – 20,788,364 |
|---|---|
| Length | 94 |
| Sequences | 3 |
| Columns | 94 |
| Reading direction | forward |
| Mean pairwise identity | 96.45 |
| Mean single sequence MFE | -28.20 |
| Consensus MFE | -26.30 |
| Energy contribution | -26.30 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.05 |
| Structure conservation index | 0.93 |
| SVM decision value | 1.78 |
| SVM RNA-class probability | 0.976675 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 20788270 94 + 27905053 UGCCCACAUUGAUAAGGGUCACAAAUUAAAUGACCCGUCGUGAGCACUGAGAGAAAUUUCGAGUGUCUGGCAGUGCAUAUUAAGGCAAACAUUU ((((....((((((.((((((.........))))))...((.((((((((((....)))).)))).)).))......))))))))))....... ( -28.60) >DroSec_CAF1 4082 94 + 1 UGCCCACAUUGAUAAGGGUCACAAAUUAAAUGACCCGUCGAGAGCACUGAGAGAAAUUUCAAGUGACUGGCAGUGCAUAUUAAGGCAAACAUUC ((((....(((((..((((((.........)))))))))))..((((((..((..((.....))..))..)))))).......))))....... ( -27.40) >DroSim_CAF1 4037 94 + 1 UGCCCACAUUGAUAAGGGUCACAAAUUAAAUGACCCGACGUGAGCACUGAGAGAAAUUUCAAGUGACUGGCAGUGCAUAUUAAGGCAAACAUUU ((((....((((((.((((((.........))))))...((.((((((((((....)))).)))).)).))......))))))))))....... ( -28.60) >consensus UGCCCACAUUGAUAAGGGUCACAAAUUAAAUGACCCGUCGUGAGCACUGAGAGAAAUUUCAAGUGACUGGCAGUGCAUAUUAAGGCAAACAUUU ((((....((((((.((((((.........)))))).......((((((..((..((.....))..))..)))))).))))))))))....... (-26.30 = -26.30 + 0.00)


| Location | 20,788,270 – 20,788,364 |
|---|---|
| Length | 94 |
| Sequences | 3 |
| Columns | 94 |
| Reading direction | reverse |
| Mean pairwise identity | 96.45 |
| Mean single sequence MFE | -25.91 |
| Consensus MFE | -25.52 |
| Energy contribution | -25.52 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.56 |
| Structure conservation index | 0.98 |
| SVM decision value | 1.86 |
| SVM RNA-class probability | 0.980176 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 20788270 94 - 27905053 AAAUGUUUGCCUUAAUAUGCACUGCCAGACACUCGAAAUUUCUCUCAGUGCUCACGACGGGUCAUUUAAUUUGUGACCCUUAUCAAUGUGGGCA .......(((((..((((((((((..(((...........)))..)))))).......(((((((.......)))))))......))))))))) ( -26.60) >DroSec_CAF1 4082 94 - 1 GAAUGUUUGCCUUAAUAUGCACUGCCAGUCACUUGAAAUUUCUCUCAGUGCUCUCGACGGGUCAUUUAAUUUGUGACCCUUAUCAAUGUGGGCA .......(((((..((((((((((..((.............))..)))))).......(((((((.......)))))))......))))))))) ( -25.32) >DroSim_CAF1 4037 94 - 1 AAAUGUUUGCCUUAAUAUGCACUGCCAGUCACUUGAAAUUUCUCUCAGUGCUCACGUCGGGUCAUUUAAUUUGUGACCCUUAUCAAUGUGGGCA .......((((.......((((((..((.............))..)))))).(((((.(((((((.......)))))))......))))))))) ( -25.82) >consensus AAAUGUUUGCCUUAAUAUGCACUGCCAGUCACUUGAAAUUUCUCUCAGUGCUCACGACGGGUCAUUUAAUUUGUGACCCUUAUCAAUGUGGGCA .......(((((..((((((((((..((.............))..)))))).......(((((((.......)))))))......))))))))) (-25.52 = -25.52 + -0.00)



| Location | 20,788,326 – 20,788,444 |
|---|---|
| Length | 118 |
| Sequences | 3 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 93.22 |
| Mean single sequence MFE | -28.00 |
| Consensus MFE | -21.76 |
| Energy contribution | -22.43 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.27 |
| Structure conservation index | 0.78 |
| SVM decision value | -0.04 |
| SVM RNA-class probability | 0.512517 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 20788326 118 + 27905053 UUUCGAGUGUCUGGCAGUGCAUAUUAAGGCAAACAUUUGUAUUAAAUGCUUAUCAUUUGUUAUUAAGUGUCAAGUCGGACAGUUAAGUGCUAUUAUUAAGGCACUAAAUGAUCUAAAA ....((.((((((((...((((.((((.((((....)))).))))))))....((((((....))))))....)))))))).)).((((((........))))))............. ( -29.70) >DroSec_CAF1 4138 118 + 1 UUUCAAGUGACUGGCAGUGCAUAUUAAGGCAAACAUUCGUUUUAAAUGCUAAUCAUUUGUUAUUAACUGUCAUGUCUGACAGUUAAGAGCUAUUAUUAAGGUACUAAAUGAUCUAAAA ...(((((((.(((((.......(((((((........))))))).))))).)))))))...((((((((((....))))))))))............((((........)))).... ( -26.10) >DroSim_CAF1 4093 118 + 1 UUUCAAGUGACUGGCAGUGCAUAUUAAGGCAAACAUUUGUUUUAAAUGCUAAUCAAUUGUUAUUAACUGUCAAGUCUGACAGUUAAGAGCUAUUAUUAGGGUACUAAAUGAUCUAAAA ...............(((((...(((((((((....)))))))))...(((((.(((.(((.((((((((((....)))))))))).))).)))))))).)))))............. ( -28.20) >consensus UUUCAAGUGACUGGCAGUGCAUAUUAAGGCAAACAUUUGUUUUAAAUGCUAAUCAUUUGUUAUUAACUGUCAAGUCUGACAGUUAAGAGCUAUUAUUAAGGUACUAAAUGAUCUAAAA ..(((((((.((......((((.(((((((((....))))))))))))).........(((.(((((((((......))))))))).)))........)).))))...)))....... (-21.76 = -22.43 + 0.67)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:53:47 2006