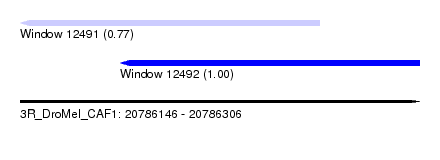
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 20,786,146 – 20,786,306 |
| Length | 160 |
| Max. P | 0.995747 |

| Location | 20,786,146 – 20,786,266 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.22 |
| Mean single sequence MFE | -39.17 |
| Consensus MFE | -38.76 |
| Energy contribution | -38.43 |
| Covariance contribution | -0.33 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.99 |
| SVM decision value | 0.53 |
| SVM RNA-class probability | 0.769687 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 20786146 120 - 27905053 CUGCCUGGCCAGCAACGCCCACAUUUGUGGGAUGCUGGAUGCAAUUCGUCUACGUCAAGCGGGCACUAAAAAUAAAAUGCCCCCGGCUGGAUUCGAAGUUGAUUCAUUCCGUUCUGUGCU ((((.(((((((((...(((((....))))).)))))((((.....))))...)))).))))((((............(((...))).(((...(((.....)))..))).....)))). ( -38.70) >DroSec_CAF1 1936 120 - 1 CUGCCUGGCCAGCAACGCCUACAUUUGUGGGAUGUUGGAUGCAAUUCGUCUACGUCAGGCGGGCACUAAAAAUAAAAUGCCCCCGGCUGGAUUCGAAGUUGAUUCAUUCCAUUCUGUUCU ..(((((((...((((((((((....))))).)))))((((.....))))...)))))))(((((............)))))..(..((((...(((.....)))..))))..)...... ( -38.30) >DroSim_CAF1 1922 120 - 1 CUGCCUGGCCAGCAACGCCUACAUUUGUGGGAUGCUGGAUGCAAUUCGUCUACGUCAGGCGGGCACUAAAAAUAAAAUGCCCCCGGCUGGAUUCGAAGUUGAUUCAUUCCAUUCUGUUCU ..((((((((((((...(((((....))))).)))))((((.....))))...)))))))(((((............)))))..(..((((...(((.....)))..))))..)...... ( -40.50) >consensus CUGCCUGGCCAGCAACGCCUACAUUUGUGGGAUGCUGGAUGCAAUUCGUCUACGUCAGGCGGGCACUAAAAAUAAAAUGCCCCCGGCUGGAUUCGAAGUUGAUUCAUUCCAUUCUGUUCU ..((((((((((((...(((((....))))).)))))((((.....))))...)))))))(((((............)))))..(..((((...(((.....)))..))))..)...... (-38.76 = -38.43 + -0.33)


| Location | 20,786,186 – 20,786,306 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.10 |
| Mean single sequence MFE | -42.73 |
| Consensus MFE | -42.93 |
| Energy contribution | -42.60 |
| Covariance contribution | -0.33 |
| Combinations/Pair | 1.08 |
| Mean z-score | -3.33 |
| Structure conservation index | 1.00 |
| SVM decision value | 2.61 |
| SVM RNA-class probability | 0.995747 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 20786186 120 - 27905053 GUGAUCUUGGCUAUUUUUUUUUUAAUUGCCUCGACCGUGUCUGCCUGGCCAGCAACGCCCACAUUUGUGGGAUGCUGGAUGCAAUUCGUCUACGUCAAGCGGGCACUAAAAAUAAAAUGC ..(.((..(((.(((........))).)))..)).)((((((((.(((((((((...(((((....))))).)))))((((.....))))...)))).)))))))).............. ( -40.90) >DroSec_CAF1 1976 119 - 1 GUGAUCUUGGCUAUUUUU-UUUUAAUUGCUUGGACCGUGUCUGCCUGGCCAGCAACGCCUACAUUUGUGGGAUGUUGGAUGCAAUUCGUCUACGUCAGGCGGGCACUAAAAAUAAAAUGC ..(.(((.(((.(((...-....))).))).))).)(((((((((((((...((((((((((....))))).)))))((((.....))))...))))))))))))).............. ( -42.30) >DroSim_CAF1 1962 118 - 1 GUGAUCUUGGCUAUUUU--UUUUAAUUGCUUGGACCGUGUCUGCCUGGCCAGCAACGCCUACAUUUGUGGGAUGCUGGAUGCAAUUCGUCUACGUCAGGCGGGCACUAAAAAUAAAAUGC ..(.(((.(((.(((..--....))).))).))).)((((((((((((((((((...(((((....))))).)))))((((.....))))...))))))))))))).............. ( -45.00) >consensus GUGAUCUUGGCUAUUUUU_UUUUAAUUGCUUGGACCGUGUCUGCCUGGCCAGCAACGCCUACAUUUGUGGGAUGCUGGAUGCAAUUCGUCUACGUCAGGCGGGCACUAAAAAUAAAAUGC ..(.((..(((.(((........))).)))..)).)((((((((((((((((((...(((((....))))).)))))((((.....))))...))))))))))))).............. (-42.93 = -42.60 + -0.33)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:53:39 2006