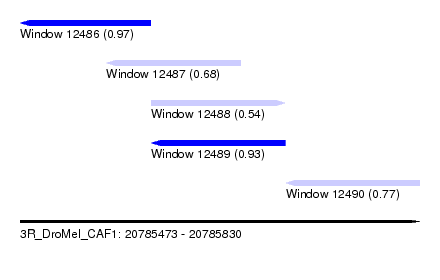
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 20,785,473 – 20,785,830 |
| Length | 357 |
| Max. P | 0.969285 |

| Location | 20,785,473 – 20,785,590 |
|---|---|
| Length | 117 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.38 |
| Mean single sequence MFE | -31.31 |
| Consensus MFE | -29.99 |
| Energy contribution | -29.77 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.48 |
| Structure conservation index | 0.96 |
| SVM decision value | 1.64 |
| SVM RNA-class probability | 0.969285 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 20785473 117 - 27905053 AUUUGCUCUGGGCCCAGGCAAAAAUGUCAGCUGUGUGGCGAGCAGCUCCCCACUUUCCGGGCACACCUC---CCCCAACCCCCUCUUUACAGGCCAAACAUACAAAUUAGAGCAAAUUCA (((((((((((((((.((((....))))......((((.(((...))).)))).....)))).......---.........(((......))).............)))))))))))... ( -31.80) >DroSec_CAF1 1260 116 - 1 AUUUGCUCUGAGCCCAGGCAAAAAUGUCAGCUGUGUGGCGAGCAGCUCCCCACUUUCCGGGCACACCUC---CCCC-CCUCCCGCUUUACAGACCAAACAUACAAAUUAGAGCAAAUACA (((((((((((((....))....((((...((((((((.(((...))).))))....((((........---....-...))))....)))).....)))).....)))))))))))... ( -31.66) >DroSim_CAF1 1243 119 - 1 AUUUGCUCUGGGCCCAGGCAAAAAUGUCAGCUGUGUGGCGAGCAGCUCCCCACUUUCCGGGCACACCUCCUUCCCC-CCUCCCGCUUUACAGACCAAACAUACAAAUUAGAGCAAAUACA (((((((((((((....))....((((...((((((((.(((...))).))))....((((...............-...))))....)))).....)))).....)))))))))))... ( -30.47) >consensus AUUUGCUCUGGGCCCAGGCAAAAAUGUCAGCUGUGUGGCGAGCAGCUCCCCACUUUCCGGGCACACCUC___CCCC_CCUCCCGCUUUACAGACCAAACAUACAAAUUAGAGCAAAUACA (((((((((((((((.((((....))))......((((.(((...))).)))).....)))).............................(......).......)))))))))))... (-29.99 = -29.77 + -0.22)

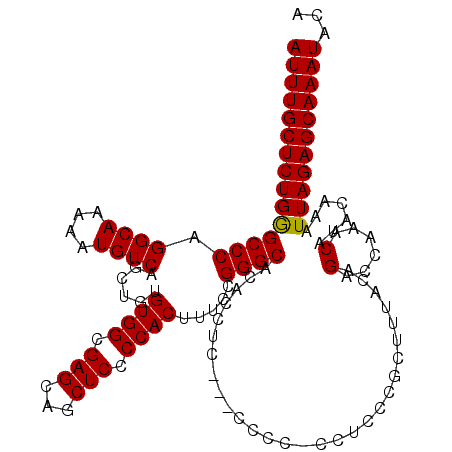
| Location | 20,785,550 – 20,785,670 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.67 |
| Mean single sequence MFE | -47.60 |
| Consensus MFE | -44.52 |
| Energy contribution | -44.85 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.94 |
| SVM decision value | 0.30 |
| SVM RNA-class probability | 0.676766 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 20785550 120 - 27905053 GGCAGCGAAUGAGCGAGCGAGUGCGACUGACUGCUCGCACUUAGACGCGUUCUAAUUAGCCAUGCCAGGCACAUUUUGGUAUUUGCUCUGGGCCCAGGCAAAAAUGUCAGCUGUGUGGCG ((((((....(((((..((((((((((.....).)))))))).)...)))))......))..)))).(.(((((.(((..((((((.(((....))))))..)))..)))..))))).). ( -43.60) >DroSec_CAF1 1336 120 - 1 GGCGACGAGUGAGCGAGCGAGUGCGACUGACUGCUCGCACUUAGCCGCGUUCUAAUUAGCCAUGCCAGGCACAGUUUGGUAUUUGCUCUGAGCCCAGGCAAAAAUGUCAGCUGUGUGGCG (((....((...(((.(((((((((((.....).)))))))).)))))...)).....)))..(((..((((((((.(..((((((.(((....))))))..)))..)))))))))))). ( -48.90) >DroSim_CAF1 1322 120 - 1 GGCGACGAGUGAGCGAGCGAGUGCGACUGACUGCUCGCACUUAGCCGCGUUCUAAUUAGCCAUGCCAGGCACAGUUUGGUAUUUGCUCUGGGCCCAGGCAAAAAUGUCAGCUGUGUGGCG (((....((...(((.(((((((((((.....).)))))))).)))))...)).....)))..(((..((((((((.(..((((((.(((....))))))..)))..)))))))))))). ( -50.30) >consensus GGCGACGAGUGAGCGAGCGAGUGCGACUGACUGCUCGCACUUAGCCGCGUUCUAAUUAGCCAUGCCAGGCACAGUUUGGUAUUUGCUCUGGGCCCAGGCAAAAAUGUCAGCUGUGUGGCG (((.......(((((.(((((((((((.....).)))))))).).).)))))......)))..(((..((((((((.(..((((((.(((....))))))..)))..)))))))))))). (-44.52 = -44.85 + 0.33)

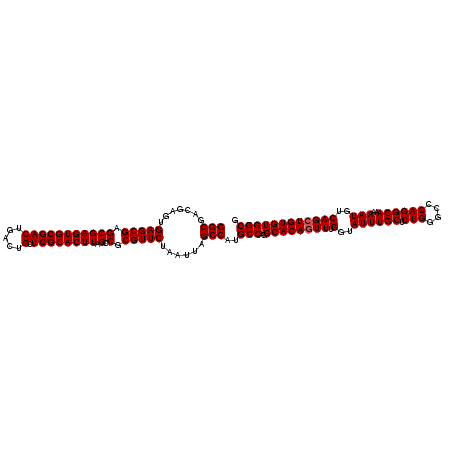
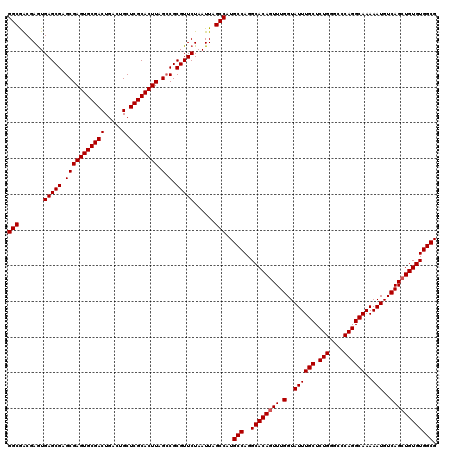
| Location | 20,785,590 – 20,785,710 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.11 |
| Mean single sequence MFE | -39.37 |
| Consensus MFE | -37.59 |
| Energy contribution | -36.93 |
| Covariance contribution | -0.66 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.95 |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.544939 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 20785590 120 + 27905053 ACCAAAAUGUGCCUGGCAUGGCUAAUUAGAACGCGUCUAAGUGCGAGCAGUCAGUCGCACUCGCUCGCUCAUUCGCUGCCACGCAUGCGCAGAUACAUUUGUAUCUUUGUAUCUUCGGGC ..........(((((((.((((...(((((.....)))))(.((((((((((....).))).)))))).).......)))).))(((((.((((((....)))))).)))))...))))) ( -40.90) >DroSec_CAF1 1376 120 + 1 ACCAAACUGUGCCUGGCAUGGCUAAUUAGAACGCGGCUAAGUGCGAGCAGUCAGUCGCACUCGCUCGCUCACUCGUCGCCACGCAUGCGCAGAUACAUUUGUAUCUUCGUAUCUUCCGGC .((.....((((.((((.(((((...........))))).(.((((((((((....).))).)))))).).......)))).))))(((.((((((....)))))).))).......)). ( -39.40) >DroSim_CAF1 1362 120 + 1 ACCAAACUGUGCCUGGCAUGGCUAAUUAGAACGCGGCUAAGUGCGAGCAGUCAGUCGCACUCGCUCGCUCACUCGUCGCCACGCAUGCGCAGAUACAUUUGUAUCUUUGUAUCUUCCGGC .((.....((((.((((.(((((...........))))).(.((((((((((....).))).)))))).).......)))).))))(((.((((((....)))))).))).......)). ( -37.80) >consensus ACCAAACUGUGCCUGGCAUGGCUAAUUAGAACGCGGCUAAGUGCGAGCAGUCAGUCGCACUCGCUCGCUCACUCGUCGCCACGCAUGCGCAGAUACAUUUGUAUCUUUGUAUCUUCCGGC .((.....((((.((((..(((...((((.......))))(.((((((((((....).))).)))))).)....))))))).))))(((.((((((....)))))).))).......)). (-37.59 = -36.93 + -0.66)

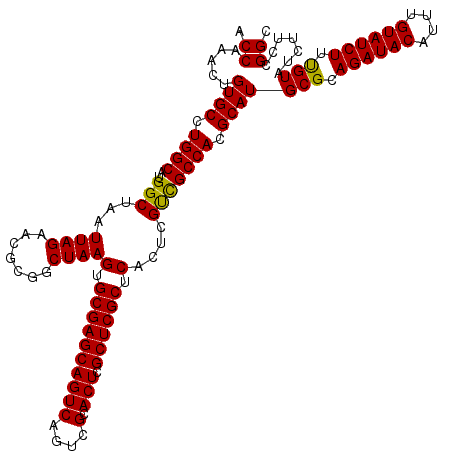
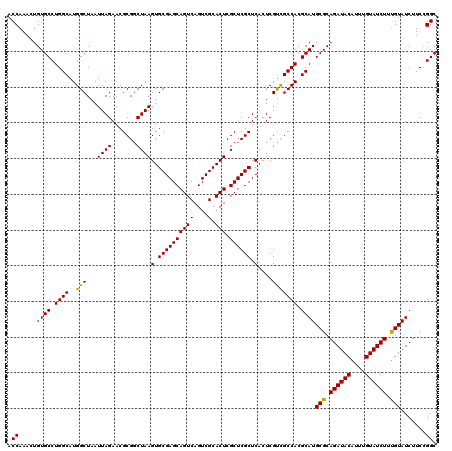
| Location | 20,785,590 – 20,785,710 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.11 |
| Mean single sequence MFE | -45.77 |
| Consensus MFE | -42.59 |
| Energy contribution | -42.92 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.42 |
| Structure conservation index | 0.93 |
| SVM decision value | 1.20 |
| SVM RNA-class probability | 0.929780 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 20785590 120 - 27905053 GCCCGAAGAUACAAAGAUACAAAUGUAUCUGCGCAUGCGUGGCAGCGAAUGAGCGAGCGAGUGCGACUGACUGCUCGCACUUAGACGCGUUCUAAUUAGCCAUGCCAGGCACAUUUUGGU ..((((((......((((((....))))))(.((..(((((((.......(((((..((((((((((.....).)))))))).)...)))))......)))))))...)).).)))))). ( -43.02) >DroSec_CAF1 1376 120 - 1 GCCGGAAGAUACGAAGAUACAAAUGUAUCUGCGCAUGCGUGGCGACGAGUGAGCGAGCGAGUGCGACUGACUGCUCGCACUUAGCCGCGUUCUAAUUAGCCAUGCCAGGCACAGUUUGGU ((((((......(.((((((....)))))).)((..(((((((....((...(((.(((((((((((.....).)))))))).)))))...)).....)))))))...))....)))))) ( -47.30) >DroSim_CAF1 1362 120 - 1 GCCGGAAGAUACAAAGAUACAAAUGUAUCUGCGCAUGCGUGGCGACGAGUGAGCGAGCGAGUGCGACUGACUGCUCGCACUUAGCCGCGUUCUAAUUAGCCAUGCCAGGCACAGUUUGGU ((((((........((((((....))))))(.((..(((((((....((...(((.(((((((((((.....).)))))))).)))))...)).....)))))))...)).)..)))))) ( -47.00) >consensus GCCGGAAGAUACAAAGAUACAAAUGUAUCUGCGCAUGCGUGGCGACGAGUGAGCGAGCGAGUGCGACUGACUGCUCGCACUUAGCCGCGUUCUAAUUAGCCAUGCCAGGCACAGUUUGGU ((((((........((((((....))))))(.((..(((((((.......(((((.(((((((((((.....).)))))))).).).)))))......)))))))...)).)..)))))) (-42.59 = -42.92 + 0.33)

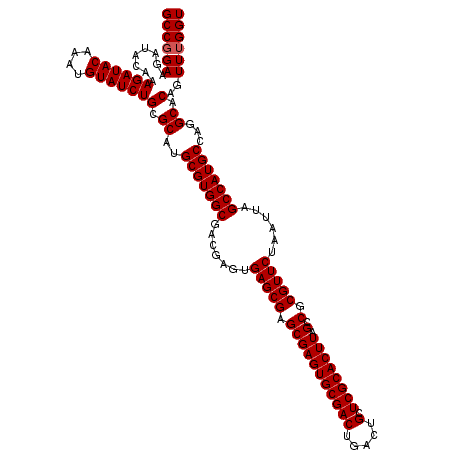

| Location | 20,785,710 – 20,785,830 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 99.44 |
| Mean single sequence MFE | -31.17 |
| Consensus MFE | -31.10 |
| Energy contribution | -31.10 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.68 |
| Structure conservation index | 1.00 |
| SVM decision value | 0.53 |
| SVM RNA-class probability | 0.772747 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 20785710 120 - 27905053 GACAAAUGUUGUCAAGCAUUUGGCGAAAAAUUGGCAAAUAGAUUCGCGCAUAAGGGGCCGCACAAAACAAAGAACAUUCGCCAAGUGCGCGUGGGUCAAGAAAGAAGUCAAGUCAAGUCA ..((((((((....))))))))..((....(((((.....(((((((((....(....)((((.....................)))))))))))))..((......))..))))).)). ( -31.10) >DroSec_CAF1 1496 120 - 1 GACAAAUGUUGUCAAGCAUUUGGCGAAAAAUUGGCAAAUAGAUUCGCGCAUAAGGGGCCGCACAAAACAAAGAACGUUCGCCAAGUGCGCGUGGGUCAAGAAAGAAGUCAAGUCAAGUCA ..((((((((....))))))))..((....(((((.....(((((((((.((...(((.(.((............)).))))...)).)))))))))..((......))..))))).)). ( -31.20) >DroSim_CAF1 1482 120 - 1 GACAAAUGUUGUCAAGCAUUUGGCGAAAAAUUGGCAAAUAGAUUCGCGCAUAAGGGGCCGCACAAAACAAAGAACGUUCGCCAAGUGCGCGUGGGUCAAGAAAGAAGUCAAGUCAAGUCA ..((((((((....))))))))..((....(((((.....(((((((((.((...(((.(.((............)).))))...)).)))))))))..((......))..))))).)). ( -31.20) >consensus GACAAAUGUUGUCAAGCAUUUGGCGAAAAAUUGGCAAAUAGAUUCGCGCAUAAGGGGCCGCACAAAACAAAGAACGUUCGCCAAGUGCGCGUGGGUCAAGAAAGAAGUCAAGUCAAGUCA ..((((((((....))))))))..((....(((((.....(((((((((....(....)((((.....................)))))))))))))..((......))..))))).)). (-31.10 = -31.10 + -0.00)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:53:37 2006