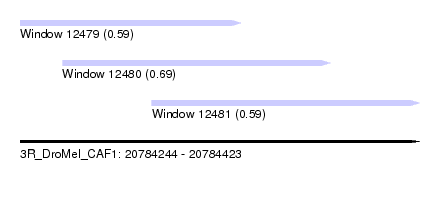
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 20,784,244 – 20,784,423 |
| Length | 179 |
| Max. P | 0.686597 |

| Location | 20,784,244 – 20,784,343 |
|---|---|
| Length | 99 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.67 |
| Mean single sequence MFE | -31.23 |
| Consensus MFE | -27.52 |
| Energy contribution | -28.93 |
| Covariance contribution | 1.42 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.591071 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 20784244 99 + 27905053 ---------------------UCACUCUCGAAGCUCCUUUCCCGAUGUCAUUAAAGGCUUAAGGAAUUCGAAUUUGAAUAGCCGUGAACUGCAGGCGACUGCACUCGGUAUCAUCUUCCC ---------------------(((...(((((..(((((..((............))...))))).)))))...)))...((((.(...(((((....)))))).))))........... ( -25.90) >DroSec_CAF1 1 120 + 1 CUAAUGGAUGUGCUCGUAUAUCCAGACUCGAAGCUCCUUUCCCGAUGUCAUUAAAGGCUUAAGGAAUUCGAAUUUGAAUAGCCGUGAACUGCAGGCGACUGCACUCGGUAUCAUCUUCCC ....((((((((....))))))))...(((((..(((((..((............))...))))).)))))...(((...((((.(...(((((....)))))).)))).)))....... ( -33.90) >DroSim_CAF1 1 120 + 1 CUAAUGGAUGUGCUCGUAUAUCCAGACUCGAAGCUCCUUUCCCGAUGUCAUUAAAGGCUUAAGGAAUUCGAAUUUGAAUAGCCGUGAACUGCAGGCGACUGCACUCGGUAUCAUCUUCCC ....((((((((....))))))))...(((((..(((((..((............))...))))).)))))...(((...((((.(...(((((....)))))).)))).)))....... ( -33.90) >consensus CUAAUGGAUGUGCUCGUAUAUCCAGACUCGAAGCUCCUUUCCCGAUGUCAUUAAAGGCUUAAGGAAUUCGAAUUUGAAUAGCCGUGAACUGCAGGCGACUGCACUCGGUAUCAUCUUCCC ....((((((((....))))))))...(((((..(((((..((............))...))))).)))))...(((...((((.(...(((((....)))))).)))).)))....... (-27.52 = -28.93 + 1.42)

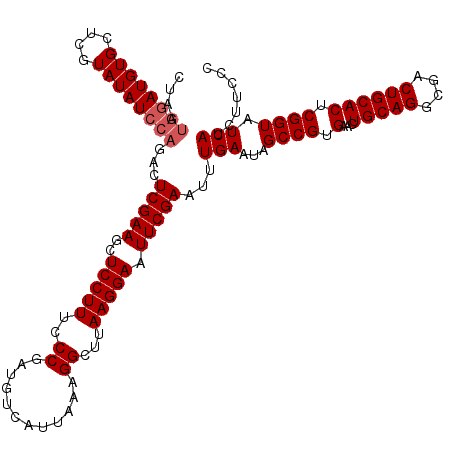
| Location | 20,784,263 – 20,784,383 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 100.00 |
| Mean single sequence MFE | -27.70 |
| Consensus MFE | -27.70 |
| Energy contribution | -27.70 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.53 |
| Structure conservation index | 1.00 |
| SVM decision value | 0.32 |
| SVM RNA-class probability | 0.686597 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 20784263 120 + 27905053 CCCGAUGUCAUUAAAGGCUUAAGGAAUUCGAAUUUGAAUAGCCGUGAACUGCAGGCGACUGCACUCGGUAUCAUCUUCCCACUACCAUUCCCGGCACUCAAUCCGUAUCCAAAUCCUCCU ...((((((......)))....(((...((...((((...((((.(((.(((((....)))))...((((............)))).))).))))..))))..))..)))..)))..... ( -27.70) >DroSec_CAF1 41 120 + 1 CCCGAUGUCAUUAAAGGCUUAAGGAAUUCGAAUUUGAAUAGCCGUGAACUGCAGGCGACUGCACUCGGUAUCAUCUUCCCACUACCAUUCCCGGCACUCAAUCCGUAUCCAAAUCCUCCU ...((((((......)))....(((...((...((((...((((.(((.(((((....)))))...((((............)))).))).))))..))))..))..)))..)))..... ( -27.70) >DroSim_CAF1 41 120 + 1 CCCGAUGUCAUUAAAGGCUUAAGGAAUUCGAAUUUGAAUAGCCGUGAACUGCAGGCGACUGCACUCGGUAUCAUCUUCCCACUACCAUUCCCGGCACUCAAUCCGUAUCCAAAUCCUCCU ...((((((......)))....(((...((...((((...((((.(((.(((((....)))))...((((............)))).))).))))..))))..))..)))..)))..... ( -27.70) >consensus CCCGAUGUCAUUAAAGGCUUAAGGAAUUCGAAUUUGAAUAGCCGUGAACUGCAGGCGACUGCACUCGGUAUCAUCUUCCCACUACCAUUCCCGGCACUCAAUCCGUAUCCAAAUCCUCCU ...((((((......)))....(((...((...((((...((((.(((.(((((....)))))...((((............)))).))).))))..))))..))..)))..)))..... (-27.70 = -27.70 + -0.00)

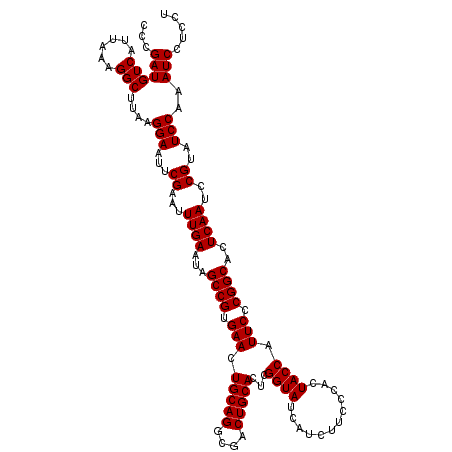
| Location | 20,784,303 – 20,784,423 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.32 |
| Mean single sequence MFE | -34.97 |
| Consensus MFE | -34.10 |
| Energy contribution | -34.43 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.98 |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.591809 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 20784303 120 + 27905053 GCCGUGAACUGCAGGCGACUGCACUCGGUAUCAUCUUCCCACUACCAUUCCCGGCACUCAAUCCGUAUCCAAAUCCUCCUGAAGUGCCGCCGUCAAAAGUUCGGGAUGAUGGGCCAUCUU ((((.(...(((((....)))))).)))).(((((.(((((((........(((((((((...................)).)))))))........)))..)))).)))))........ ( -35.40) >DroSec_CAF1 81 118 + 1 GCCGUGAACUGCAGGCGACUGCACUCGGUAUCAUCUUCCCACUACCAUUCCCGGCACUCAAUCCGUAUCCAAAUCCUCCUGAAGUGCCGCCGUCA--AUUUCGGGAUGAUGGGCCAUCUU ((((.(...(((((....)))))).)))).(((((.((((...........(((((((((...................)).)))))))......--.....)))).)))))........ ( -34.76) >DroSim_CAF1 81 118 + 1 GCCGUGAACUGCAGGCGACUGCACUCGGUAUCAUCUUCCCACUACCAUUCCCGGCACUCAAUCCGUAUCCAAAUCCUCCUGAAGUGCCGCCGUCA--AGUUCGGGAUGAUGGGCCAUCUU ((((.(...(((((....)))))).)))).(((((.((((...........(((((((((...................)).)))))))......--.....)))).)))))........ ( -34.76) >consensus GCCGUGAACUGCAGGCGACUGCACUCGGUAUCAUCUUCCCACUACCAUUCCCGGCACUCAAUCCGUAUCCAAAUCCUCCUGAAGUGCCGCCGUCA__AGUUCGGGAUGAUGGGCCAUCUU ((((.(...(((((....)))))).)))).(((((.(((((((........(((((((((...................)).)))))))........)))..)))).)))))........ (-34.10 = -34.43 + 0.33)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:53:23 2006