| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 20,773,895 – 20,774,014 |
| Length | 119 |
| Max. P | 0.603987 |

| Location | 20,773,895 – 20,774,014 |
|---|---|
| Length | 119 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.86 |
| Mean single sequence MFE | -39.17 |
| Consensus MFE | -27.41 |
| Energy contribution | -26.97 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.35 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.603987 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
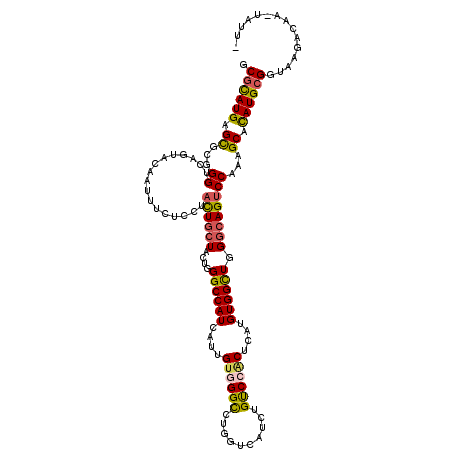
>3R_DroMel_CAF1 20773895 119 + 27905053 GCGCAUGAGCGCUGGUCAGUACAAUUUCUCUUACUGCUACUGGGCCAUCAUAGUCGGCCUGGUCAUCUGCCCGCUCAUGUGGCUGGGAAGUCCAAAGCACAUGCGGUAAGAUAA-AUUUC ((((....))))..........(((((.(((((((((.((..((((.........))))..))....(((((((....))))...((....))...)))...))))))))).))-))).. ( -39.80) >DroVir_CAF1 45827 118 + 1 CCGCAUGAGCGCUGGCCAAUACAGUUUCUCCUACUGCUAUUGGGCCAUUGUUGUGGGUCUCGUUAUCUGUCCACUCAUGUGGCUUGGCAGUCCCAAGCACAUGCGGUAAUACGA-UCUU- (((((((.((..(((((((((((((.......)))).)))).)))))......((((.((.((((...(.((((....))))).)))))).)))).)).)))))))........-....- ( -44.80) >DroGri_CAF1 49797 117 + 1 GCGCAUGAGUGCGGGUCAUUAUAAUUUCUCCUAUUGCUAUUGGGCCAUCCUUGUGGGCAUAGUCAUCUGUCCACUCAUGUGGUUGGCCAGUCCGAAGCAUAUGCGGUAAG-CAA-UAUU- ((((....))))...................(((((((....(((((.((..(((((((........)))))))......)).)))))........((....))....))-)))-))..- ( -35.00) >DroWil_CAF1 19990 119 + 1 GCGUAUGAGUGCAGGUCAUUACAAUUUCUCCUAUUGCUACUGGGCCAUAAUUGUGGGUCUGGUGAUUUGCCCCCUCAUGUGGCUGGGCAGUCCAAAGCAUAUGAGGUAAGAUUG-GAAUC (((......))).........((((((..(((..((((((..((((.........))))..))(..((((((((......))..))))))..)..))))....)))..))))))-..... ( -34.80) >DroMoj_CAF1 37582 118 + 1 ACGCAUGAGCGCCGGCCUCUAUAAUUUCUCCUACUGCUACUGGGCCAUUAUUGUGGGCCUGGUCAUCUGUCCACUCAUGUGGCUAGGCAGCCCCAAGCACAUGCGGUUAGACAA-UGUU- .((((((.((...(((..(((((((....((((.......)))).....)))))))((((((((((.((......)).)))))))))).)))....)).)))))).........-....- ( -41.70) >DroAna_CAF1 15736 120 + 1 GCGCAUGAGCGGGGGUCAGUACAACUUCUCCUACUGCUACUGGGCCAUCAUAGUCGGUCUGGUAAUCUGUCCGCUCAUGUGGCUGGGCAGUCCGAAGCAUAUGCGGUAAGAAAUUGGUUC .((((((.(((((((..((.....)).)))))((((((((..((((.........))))..)))..(((.((((....)))).)))))))).....)).))))))............... ( -38.90) >consensus GCGCAUGAGCGCGGGUCAGUACAAUUUCUCCUACUGCUACUGGGCCAUCAUUGUGGGCCUGGUCAUCUGUCCACUCAUGUGGCUGGGCAGUCCAAAGCACAUGCGGUAAGACAA_UAUU_ .((((((.((...((.................((((((....((((((....((((((..........))))))....)))))).))))))))...)).))))))............... (-27.41 = -26.97 + -0.44)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:53:14 2006