
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 20,762,626 – 20,762,801 |
| Length | 175 |
| Max. P | 0.669230 |

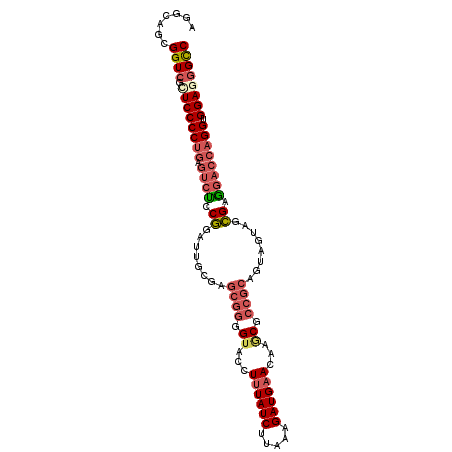
| Location | 20,762,626 – 20,762,727 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 83.23 |
| Mean single sequence MFE | -38.82 |
| Consensus MFE | -27.78 |
| Energy contribution | -29.01 |
| Covariance contribution | 1.23 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.29 |
| SVM RNA-class probability | 0.669230 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 20762626 101 - 27905053 AGGCAGCGGUCGCUCCCCUGAGUCUUCGGAUUGCGAGCGGGGUACCUUUAUCUUAAAGAUGCACAAGCGCCGCAGUAGUAGCGAGGACCAGGUGGAGGGCC .......((((.((((((((.(((((((.(((((..((((.((.....((((.....)))).....)).)))).)))))..))))))))))).)))))))) ( -46.40) >DroSec_CAF1 5031 101 - 1 AGGAAGCGGUCGCUCCCCUGAGUCUCCGGAUUGCGAGCGGGGUACUUUUAUCUUAAAGAUGAACAAGCGCCGCAGUAGUAGCGAAGACCAGGUGGAGGGCC .......((((.((((((((.((((.((.(((((..((((.((...((((((.....))))))...)).)))).)))))..)).)))))))).)))))))) ( -45.00) >DroSim_CAF1 5789 101 - 1 AGGAAGCGGUCGCUCCCCUGAGUCUCCGGAUUGCGAGCGGGGUACCUUUAUCUUAAAGAUGAACAAACGCCGCAGUAGUAGCGAAGACCAGGUGGAGGGCC .......((((.((((((((.((((.((.(((((..((((.((...((((((.....))))))...)).)))).)))))..)).)))))))).)))))))) ( -44.70) >DroEre_CAF1 6560 101 - 1 AGGCAGUGGUCGGUCCCCUGAGUCUCCGGAUCGCGAGCGGGGUACGUUUAUCUUAAAGAUGAACAAACGCCGCAGUAGUAGCGAGGACCAGGUGGAGGGCC .......((((..(((((((.(((..((.((..(..((((.((..(((((((.....)))))))..)).)))).)..))..))..))))))).))).)))) ( -41.00) >DroWil_CAF1 8183 79 - 1 -------GGUACAUCCCC------CCCAGAU---------GGAACCUUUAUCCUGAAGAUGAACAAGCGCCGCAGCAGCUCUGAGGAUCAGGUGGAGGGUC -------.......((((------(((....---------)).((((..(((((..(((.(.....((......))..)))).))))).)))))).))).. ( -18.60) >DroYak_CAF1 4758 101 - 1 AGGCAGUGGUCGUUCCCCUGAGUCUCCGGAUCGAGAGCGGGGUACGUUUAUCUUAAAGAUGAACAAGCGCCGCAGUAGUAGCGAGGACCAGGUGGAGGGUC .......(..(.((((((((.(((..((...(....((((.((..(((((((.....)))))))..)).))))....)...))..))))))).)))))..) ( -37.20) >consensus AGGCAGCGGUCGCUCCCCUGAGUCUCCGGAUUGCGAGCGGGGUACCUUUAUCUUAAAGAUGAACAAGCGCCGCAGUAGUAGCGAGGACCAGGUGGAGGGCC .......((((.((((((((.((((.((........((((.((...((((((.....))))))...)).))))........)).)))))))).)))))))) (-27.78 = -29.01 + 1.23)


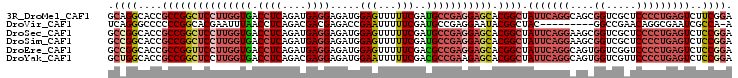
| Location | 20,762,697 – 20,762,801 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 83.72 |
| Mean single sequence MFE | -43.72 |
| Consensus MFE | -31.53 |
| Energy contribution | -33.82 |
| Covariance contribution | 2.28 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.568307 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 20762697 104 - 27905053 GCAGGCACCGCCGGCUCCUUGGUGACCUCAGAUGAGGAGAUGGAGUUUUUCGAUGCCGAGGAGCACGGCUAUUCAGGCAGCGGUCGCUCCCCUGAGUCUUCGGA (.((((...((((((((((((((..((((....))))..((.(((...))).)))))))))))).))))...(((((.(((....)))..))))))))).)... ( -45.80) >DroVir_CAF1 33944 94 - 1 UCAGGGCCCCCCGGCACGAAUUUAACCUCAGACGACGAGACCGAAUUUUUCGAUGCCGAGGAAUACGGCUAC---------GGCCGAACAGGCGAAUCGCCA-A ........((.(((((((((.((...(((.......)))....))...)))).))))).))....((((...---------.))))....((((...)))).-. ( -30.40) >DroSec_CAF1 5102 104 - 1 GCCGGCACCGCCGGCUCCUUGGUGACCUCAGAUGAGGAGAUGGAGUUUUUCGAUGCCGAGGAGCACGGCUAUUCAGGAAGCGGUCGCUCCCCUGAGUCUCCGGA .((((....((((((((((((((..((((....))))..((.(((...))).)))))))))))).)))).(((((((.(((....)))..)))))))..)))). ( -50.10) >DroSim_CAF1 5860 104 - 1 GCCGGCACCGCCGGCUCCUUGGUGACCUCAGAUGAGGAGAUGGAGUUUUUCGAUGCCGAGGAGCACGGCUAUUCAGGAAGCGGUCGCUCCCCUGAGUCUCCGGA .((((....((((((((((((((..((((....))))..((.(((...))).)))))))))))).)))).(((((((.(((....)))..)))))))..)))). ( -50.10) >DroEre_CAF1 6631 104 - 1 GCCGGCACCGCCGGUUCCUUGGUGACCUCAGAUGAGGAGAUGGAGUUUUUCGACGCCGAGGAGCACGGCUAUUCAGGCAGUGGUCGGUCCCCUGAGUCUCCGGA .((((....(((((((((((((((.((((....)))).((.........))..))))))))))).)))).(((((((....((.....)))))))))..)))). ( -46.00) >DroYak_CAF1 4829 104 - 1 GCUGGCACCGCCGGCUCCUUGGUGACCUCAGACGAGGAGAUGGAAUUUUUCGACGCCGAAGAGCACGGCUAUUCAGGCAGUGGUCGUUCCCCUGAGUCUCCGGA .((((....((((((((.((((((.((((....))))...((((....)))).)))))).)))).)))).(((((((....((.....)))))))))..)))). ( -39.90) >consensus GCCGGCACCGCCGGCUCCUUGGUGACCUCAGAUGAGGAGAUGGAGUUUUUCGAUGCCGAGGAGCACGGCUAUUCAGGCAGCGGUCGCUCCCCUGAGUCUCCGGA .((((....(((((((((((((((.((((....)))).....(((...)))..))))))))))).)))).(((((((....((.....)))))))))..)))). (-31.53 = -33.82 + 2.28)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:53:11 2006