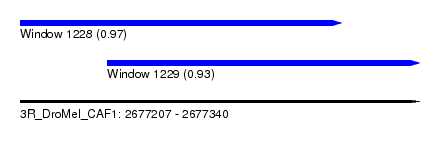
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 2,677,207 – 2,677,340 |
| Length | 133 |
| Max. P | 0.967093 |

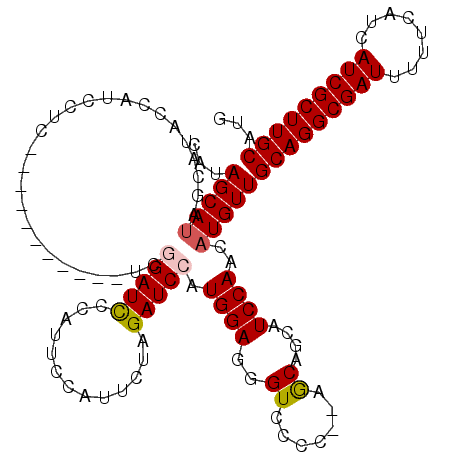
| Location | 2,677,207 – 2,677,314 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.08 |
| Mean single sequence MFE | -29.94 |
| Consensus MFE | -22.22 |
| Energy contribution | -22.42 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.68 |
| Structure conservation index | 0.74 |
| SVM decision value | 1.61 |
| SVM RNA-class probability | 0.967093 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 2677207 107 + 27905053 CAUAGCAUAGCAUAGCAUCCUC-----------UCGGAUCCCAUUCCAUUCUAGAUCCAUGGAGGGUCCCC--AGCAGCAUCCAACAUGUUGCAGGCGAUUUUUCAUCAUCGCUUGCAUG .........(((.(((......-----------..(((((.............)))))((((((((((.((--.(((((((.....))))))).)).))))))))))....))))))... ( -32.12) >DroSec_CAF1 36630 107 + 1 CAUAGCAUAGCAUACCAUCCUC-----------UCGGAUCCCAUUCCAUUCUAGAUCCAUGGAGGGUCCCC--AGCAGCAUCCAACAUGUUGCAGGCGAUUUUUCAUCAUCGCUUGCAUG ....(((.(((...........-----------..(((((.............)))))((((((((((.((--.(((((((.....))))))).)).))))))))))....))))))... ( -32.12) >DroSim_CAF1 29826 107 + 1 CAUAGCAUAGCAUACCAUCCUC-----------UCGGAUCCCAUUCCAUUCUAGAUCCAUGGAGGGUCCCC--AGCAGCAUCCAACAUGUUGCAGGCGAUUUUUCAUCAUCGCUUGCAUG ....(((.(((...........-----------..(((((.............)))))((((((((((.((--.(((((((.....))))))).)).))))))))))....))))))... ( -32.12) >DroYak_CAF1 37111 102 + 1 CAUAGCAUA-----CCAUCCGC-----------GCAGAUCCCAUUCCAUUCUAGAUCCAUGGAGGGUCCCC--AACAGCAUCCAACAUGUUGCAGGCGAUUUUUCAUCAUCGCUUGCAUG ...(((((.-----......((-----------...(((((...(((((.........))))))))))...--....)).......)))))(((((((((........)))))))))... ( -24.34) >DroAna_CAF1 24867 117 + 1 CAGAGCAACCCGUACCAUCCUCCUCCCUCCAUCCCAGAUUCCAU---AUCCCAGAUCCAUGGAGGGUCCCCUCGACAGCAUCCAACAUGUUGCAGGCGAUUUUUCAUCAUCGCUUGCAUG ....((...................(((((((....((((....---......)))).)))))))(((.....))).))...........((((((((((........)))))))))).. ( -29.00) >consensus CAUAGCAUAGCAUACCAUCCUC___________UCGGAUCCCAUUCCAUUCUAGAUCCAUGGAGGGUCCCC__AGCAGCAUCCAACAUGUUGCAGGCGAUUUUUCAUCAUCGCUUGCAUG ...(((((...........................(((((.............))))).((((..((.......))....))))..)))))(((((((((........)))))))))... (-22.22 = -22.42 + 0.20)

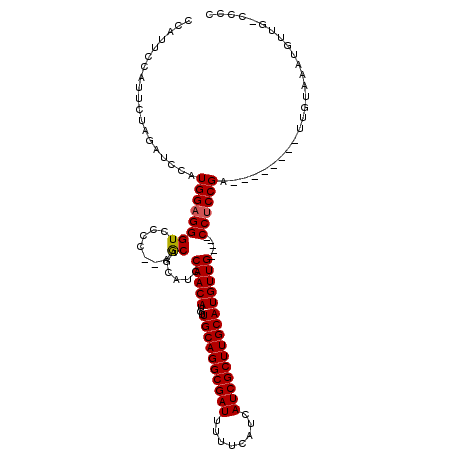
| Location | 2,677,236 – 2,677,340 |
|---|---|
| Length | 104 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.95 |
| Mean single sequence MFE | -30.68 |
| Consensus MFE | -25.20 |
| Energy contribution | -25.16 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.82 |
| SVM decision value | 1.23 |
| SVM RNA-class probability | 0.934050 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 2677236 104 + 27905053 CCAUUCCAUUCUAGAUCCAUGGAGGGUCCCC--AGCAGCAUCCAACAUGUUGCAGGCGAUUUUUCAUCAUCGCUUGCAUGUUG-----CCUCCGA--------UUGUAAAUGUUG-CCCC ......((((...((((...(((((.....(--((((((((.....))))((((((((((........)))))))))))))))-----)))))))--------))...))))...-.... ( -29.20) >DroSec_CAF1 36659 104 + 1 CCAUUCCAUUCUAGAUCCAUGGAGGGUCCCC--AGCAGCAUCCAACAUGUUGCAGGCGAUUUUUCAUCAUCGCUUGCAUGUUG-----CCUCCGA--------UUGUAAAUGUUG-CCCC ......((((...((((...(((((.....(--((((((((.....))))((((((((((........)))))))))))))))-----)))))))--------))...))))...-.... ( -29.20) >DroSim_CAF1 29855 104 + 1 CCAUUCCAUUCUAGAUCCAUGGAGGGUCCCC--AGCAGCAUCCAACAUGUUGCAGGCGAUUUUUCAUCAUCGCUUGCAUGUUG-----CCUCCGA--------UUGUAAAUGUUG-CCCC ......((((...((((...(((((.....(--((((((((.....))))((((((((((........)))))))))))))))-----)))))))--------))...))))...-.... ( -29.20) >DroYak_CAF1 37135 104 + 1 CCAUUCCAUUCUAGAUCCAUGGAGGGUCCCC--AACAGCAUCCAACAUGUUGCAGGCGAUUUUUCAUCAUCGCUUGCAUGUUG-----CCUCCGA--------GUGUAAAUGUUG-CCCC .((((.(((((..(((((.....)))))..(--((((((((.....))))((((((((((........)))))))))))))))-----.....))--------)))..))))...-.... ( -29.10) >DroAna_CAF1 24907 117 + 1 CCAU---AUCCCAGAUCCAUGGAGGGUCCCCUCGACAGCAUCCAACAUGUUGCAGGCGAUUUUUCAUCAUCGCUUGCAUGUUGUUACCCCGCCGAUGUGGAUAUUGAAGUUGUUGGUGCC ....---......(((((.....)))))...((((((((.(((((((...((((((((((........))))))))))))))).....((((....)))).....)).)))))))).... ( -36.70) >consensus CCAUUCCAUUCUAGAUCCAUGGAGGGUCCCC__AGCAGCAUCCAACAUGUUGCAGGCGAUUUUUCAUCAUCGCUUGCAUGUUG_____CCUCCGA________UUGUAAAUGUUG_CCCC ...................((((((((.......))......(((((...((((((((((........))))))))))))))).....)))))).......................... (-25.20 = -25.16 + -0.04)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:51:42 2006