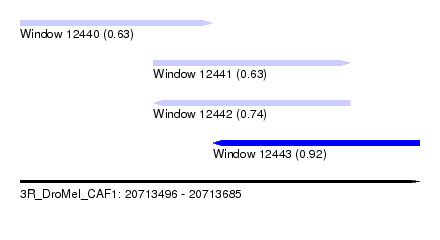
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 20,713,496 – 20,713,685 |
| Length | 189 |
| Max. P | 0.918855 |

| Location | 20,713,496 – 20,713,587 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 93 |
| Reading direction | forward |
| Mean pairwise identity | 80.81 |
| Mean single sequence MFE | -30.78 |
| Consensus MFE | -21.74 |
| Energy contribution | -23.58 |
| Covariance contribution | 1.83 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.37 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.625406 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
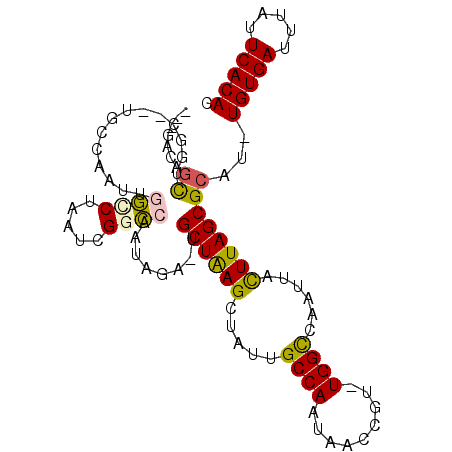
>3R_DroMel_CAF1 20713496 91 + 27905053 GUUGCAUGAUCGGUCAGAUCGGACAGAAA--CUAAAGCCGCAGCUAGCGGAUAUUUGCCUGUCAACGAGGCCGGCUGCUGUUGCUGCUGGCUG .(((..(((((.....)))))..)))...--....(((((((((.(((((......(((.(((.....))).)))..)))))))))).)))). ( -34.00) >DroSec_CAF1 9892 91 + 1 GUUGCAUGAUCGGUCAGAUCGGACAGAAA--CUAAAGCCGCAGCUAGCGGAUAUUUGCCUGUCAACGAGGCCGGCUGCUGCGGCUGCUGGCUG .(((..(((((.....)))))..)))...--....((((((((((.((((......(((.(((.....))).)))..)))))))))).)))). ( -35.70) >DroSim_CAF1 10777 91 + 1 GUUGCAUGAUCGGUCAGAUCGGACAGAAA--CUAAAGCCGCAGCUAGCGGAUAUUUGCCUGUCAACGAGGCCGGCUGCUGCGGCUGCUGGCUG .(((..(((((.....)))))..)))...--....((((((((((.((((......(((.(((.....))).)))..)))))))))).)))). ( -35.70) >DroEre_CAF1 10652 85 + 1 GUUGCAUGAUCGGUCAGAUCGGACAGAAA--CUAAAGCCGCAGCUAGCGGAUAUUUGCCUGUCAACGAGGCCGGCUGC------UGCUGGCUG (((...(((((.....)))))(((((...--.((((.((((.....))))...)))).))))))))..(((((((...------.))))))). ( -30.90) >DroYak_CAF1 10865 78 + 1 GUUGCAUGAUCGGUCAGAUCGGACAGAAA--CUAAAGCCGCAGCUAGCGGAUAUUUGCCUGUCAACGAGGCC-------------GCUGGCUG .(((..(((((.....)))))..)))...--.........((((((((((...((((........)))).))-------------)))))))) ( -26.20) >DroPer_CAF1 10216 73 + 1 GUUGCAUGAU---UCCAUUGGGACCCCGACUCCAUUGCCGU--GCAGCGGAUAUUCGCCUGUCAACGGAGUC---------------AAGCUG ...((.((((---(((.((((.(...(((.(((.((((...--)))).)))...)))..).)))).))))))---------------).)).. ( -22.20) >consensus GUUGCAUGAUCGGUCAGAUCGGACAGAAA__CUAAAGCCGCAGCUAGCGGAUAUUUGCCUGUCAACGAGGCCGGCUGC______UGCUGGCUG (((...(((((.....)))))(((((...........((((.....))))........))))))))..(((((((.((....)).))))))). (-21.74 = -23.58 + 1.83)



| Location | 20,713,559 – 20,713,652 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 83.64 |
| Mean single sequence MFE | -33.50 |
| Consensus MFE | -21.49 |
| Energy contribution | -22.05 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.57 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.628187 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 20713559 93 + 27905053 CGAG--GCCGG-CUGCUGUUGCUGCUG--------GCUGGUGUGAAUAAGUCACA-AUGCGCUAAGUAAUGGGCCAACGGUUAUUGGCAAUAGCUUAGCACUCUA .(((--(((((-(.((....)).))))--------))...(((((.....)))))-....(((((((.((..(((((......))))).)).))))))).))).. ( -37.90) >DroSec_CAF1 9955 93 + 1 CGAG--GCCGG-CUGCUGCGGCUGCUG--------GCUGGUGUGAAUAAGUCACA-GUGCGCUAAAUAAUGGGCCAACGGUUAUUGGCAAUAGCUUAGCACUCUA .(((--(((((-(.((....)).))))--------)).(.(((((.....)))))-.)..(((((.......(((((......)))))......))))).))).. ( -32.32) >DroSim_CAF1 10840 93 + 1 CGAG--GCCGG-CUGCUGCGGCUGCUG--------GCUGGUGUGAAUAAGUCACA-AUGCGCUAAGUAAUGGGCCAAAAGUUAUUGGCAAUAGCUUAGCACUCUA .(((--(((((-(.((....)).))))--------))...(((((.....)))))-....(((((((.((..(((((......))))).)).))))))).))).. ( -37.90) >DroEre_CAF1 10715 87 + 1 CGAG--GCCGG-CUGC------UGCUG--------GCUGGUGUGAAUAAAUCACA-AUGCGCUAAGUAAUGGGCCAACGGUUAUUGGCAGUAGCUUAGCACUCUA .(((--(((((-(...------.))))--------))...(((((.....)))))-....(((((((.((..(((((......))))).)).))))))).))).. ( -35.20) >DroYak_CAF1 10928 80 + 1 CGAG--GCC--------------GCUG--------GCUGGUGUGAAUAAAUCACA-AUGCGCUAAGUAAUGGGCCAACGGUUAUUGGCAAUAGCUUAGCACUCUA .(((--...--------------....--------((...(((((.....)))))-..))(((((((.((..(((((......))))).)).))))))).))).. ( -27.50) >DroAna_CAF1 10300 103 + 1 CGGGCAGCCGGCCUGCUGCUGGUACUGUUUCUAUUUCUGGUGUGAAUAAAUCACAAAUGCGCUAAGUAAUUGGCCAACGGUUAUUGGCAAUAGCUCAGC--UAAG (((((.....))))).(((..(((((((............(((((.....)))))..((.(((((....))))))))))).)))..))).((((...))--)).. ( -30.20) >consensus CGAG__GCCGG_CUGCUGC_GCUGCUG________GCUGGUGUGAAUAAAUCACA_AUGCGCUAAGUAAUGGGCCAACGGUUAUUGGCAAUAGCUUAGCACUCUA .(((...............................((...(((((.....)))))...))(((((((.((..(((((......))))).)).))))))).))).. (-21.49 = -22.05 + 0.56)
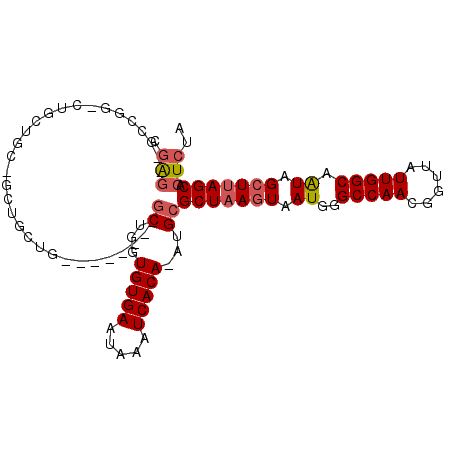

| Location | 20,713,559 – 20,713,652 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 83.64 |
| Mean single sequence MFE | -28.85 |
| Consensus MFE | -20.28 |
| Energy contribution | -20.50 |
| Covariance contribution | 0.22 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.56 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.45 |
| SVM RNA-class probability | 0.741022 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 20713559 93 - 27905053 UAGAGUGCUAAGCUAUUGCCAAUAACCGUUGGCCCAUUACUUAGCGCAU-UGUGACUUAUUCACACCAGC--------CAGCAGCAACAGCAG-CCGGC--CUCG ....((((((((..((.((((((....))))))..))..))))))))..-(((((.....)))))...((--------(.((.((....)).)-).)))--.... ( -31.00) >DroSec_CAF1 9955 93 - 1 UAGAGUGCUAAGCUAUUGCCAAUAACCGUUGGCCCAUUAUUUAGCGCAC-UGUGACUUAUUCACACCAGC--------CAGCAGCCGCAGCAG-CCGGC--CUCG ....((((((((..((.((((((....))))))..))..))))))))..-(((((.....)))))...((--------(.((.((....)).)-).)))--.... ( -28.20) >DroSim_CAF1 10840 93 - 1 UAGAGUGCUAAGCUAUUGCCAAUAACUUUUGGCCCAUUACUUAGCGCAU-UGUGACUUAUUCACACCAGC--------CAGCAGCCGCAGCAG-CCGGC--CUCG ....((((((((..((.(((((......)))))..))..))))))))..-(((((.....)))))...((--------(.((.((....)).)-).)))--.... ( -30.10) >DroEre_CAF1 10715 87 - 1 UAGAGUGCUAAGCUACUGCCAAUAACCGUUGGCCCAUUACUUAGCGCAU-UGUGAUUUAUUCACACCAGC--------CAGCA------GCAG-CCGGC--CUCG ....((((((((.....((((((....))))))......))))))))..-(((((.....)))))...((--------(.((.------...)-).)))--.... ( -29.40) >DroYak_CAF1 10928 80 - 1 UAGAGUGCUAAGCUAUUGCCAAUAACCGUUGGCCCAUUACUUAGCGCAU-UGUGAUUUAUUCACACCAGC--------CAGC--------------GGC--CUCG ....((((((((..((.((((((....))))))..))..))))))))..-(((((.....)))))...((--------(...--------------)))--.... ( -25.00) >DroAna_CAF1 10300 103 - 1 CUUA--GCUGAGCUAUUGCCAAUAACCGUUGGCCAAUUACUUAGCGCAUUUGUGAUUUAUUCACACCAGAAAUAGAAACAGUACCAGCAGCAGGCCGGCUGCCCG ....--((((((..(((((((((....))))).))))..)))))).....(((((.....))))).....................(((((......)))))... ( -29.40) >consensus UAGAGUGCUAAGCUAUUGCCAAUAACCGUUGGCCCAUUACUUAGCGCAU_UGUGACUUAUUCACACCAGC________CAGCAGC_GCAGCAG_CCGGC__CUCG ....((((((((.....((((((....))))))......))))))))...(((((.....)))))........................................ (-20.28 = -20.50 + 0.22)


| Location | 20,713,587 – 20,713,685 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 81.46 |
| Mean single sequence MFE | -32.13 |
| Consensus MFE | -17.61 |
| Energy contribution | -17.20 |
| Covariance contribution | -0.41 |
| Combinations/Pair | 1.29 |
| Mean z-score | -3.25 |
| Structure conservation index | 0.55 |
| SVM decision value | 1.12 |
| SVM RNA-class probability | 0.918855 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 20713587 98 - 27905053 ---GACAGCUGGC--UGGCAAUUGGCCUAAUCGGCCAAUAGAGUGCUAAGCUAUUGCCAAUAACCGU-UGGCCCAUUACUUAGCGCAU-UGUGACUUAUUCACAC ---..(((....)--))...(((((((.....)))))))...((((((((..((.((((((....))-))))..))..))))))))..-(((((.....))))). ( -35.30) >DroPse_CAF1 10292 98 - 1 CAGGCCAGUUGCCAAUGCCAAUUGACCCAAUCGUUG-----U-GGCUAAGCUGUUGCCAAUAACCGUUUGGUCAAUUACUUAGCGCAU-UGUGAUUUAUUCACAC ..(((.....)))((((((((((((((.(((.((((-----(-(((.........))).))))).))).)))))))).....).))))-)((((.....)))).. ( -29.80) >DroSec_CAF1 9983 98 - 1 ---GACAGCUGGC--UGGCAAUUGGCCUAAUCGGCCAAUAGAGUGCUAAGCUAUUGCCAAUAACCGU-UGGCCCAUUAUUUAGCGCAC-UGUGACUUAUUCACAC ---..(((....)--))...(((((((.....)))))))...((((((((..((.((((((....))-))))..))..))))))))..-(((((.....))))). ( -33.00) >DroEre_CAF1 10737 98 - 1 ---GACAGCUGGC--UGGCAAUUGGCCUAAUCGGCCAAUAGAGUGCUAAGCUACUGCCAAUAACCGU-UGGCCCAUUACUUAGCGCAU-UGUGAUUUAUUCACAC ---..(((....)--))...(((((((.....)))))))...((((((((.....((((((....))-))))......))))))))..-(((((.....))))). ( -36.00) >DroAna_CAF1 10339 95 - 1 -----CACCCGGC--UGCCAAUUGGUCUAAUCGGCCUGCUUA--GCUGAGCUAUUGCCAAUAACCGU-UGGCCAAUUACUUAGCGCAUUUGUGAUUUAUUCACAC -----.....(((--((((....)))......))))(((...--((((((..(((((((((....))-))).))))..)))))))))..(((((.....))))). ( -28.90) >DroPer_CAF1 10289 98 - 1 CAGGCCAGUUGCCAAUGCCAAUUGACCUAAUCGUUG-----U-GGCUAAGCUGUUGCCAAUAACCGUUUGGUCAAUUACUUAGCGCAU-UGUGAUUUAUUCACAC ..(((.....)))((((((((((((((.(((.((((-----(-(((.........))).))))).))).)))))))).....).))))-)((((.....)))).. ( -29.80) >consensus ___GACAGCUGGC__UGCCAAUUGGCCUAAUCGGCCAAUAGA_UGCUAAGCUAUUGCCAAUAACCGU_UGGCCAAUUACUUAGCGCAU_UGUGAUUUAUUCACAC .......((..............((((.....))))........((((((.....((((.........))))......))))))))...(((((.....))))). (-17.61 = -17.20 + -0.41)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:52:47 2006