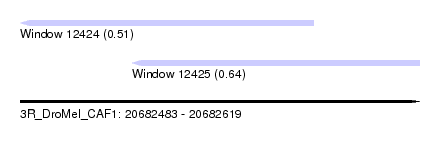
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 20,682,483 – 20,682,619 |
| Length | 136 |
| Max. P | 0.639253 |

| Location | 20,682,483 – 20,682,583 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 79.74 |
| Mean single sequence MFE | -26.55 |
| Consensus MFE | -17.40 |
| Energy contribution | -18.57 |
| Covariance contribution | 1.17 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.26 |
| Structure conservation index | 0.66 |
| SVM decision value | -0.04 |
| SVM RNA-class probability | 0.511954 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 20682483 100 - 27905053 UAAACAGGUAUCCAGCA--UAACUAAGUUGGAUGAUUCGUGUCCUCUGGAGGACAGCAUGGCUUUGCAGGACCUUUUUGCUAGCAUAGACAGCCAAAAAUAC ......((((((((((.--.......)))))))...((.((((((....)))))).....(((..((((((...)))))).)))...))..)))........ ( -26.90) >DroPse_CAF1 1673 100 - 1 CAAAGAGGUC--AAGCUGCUCAGAAAGUGCUACGACUCCUCACAAGUGAAGGCUGAAAUUUCACUGGUAGAUCUGUUUGCUGGCAUCGAUAUGGAUACAUUU ....((.(((--(.((....((((.....((((((....))...(((((((.......))))))).)))).))))...)))))).))............... ( -20.20) >DroSec_CAF1 1645 100 - 1 CAAACAGGUAUCCAGCA--UAACUAAGUUGGAUGAUUCGUGUCCUCUGGAGGACAGUAUGGCUCUGCAAGAUCUGUUUGUAAGCAUAGAUAGCCAGAAAUAC ........((((((((.--.......)))))))).(((.((((((....))))))...(((((.......((((((........)))))))))))))).... ( -27.21) >DroSim_CAF1 1661 100 - 1 UAAACAGGUAUCCAGCA--UAGCUAAGUUGGAUGAUUCGUGUCCUCUGGAGGACAGUAUGGCUCUGCAAGAUCUGUUUGUAAGCAUAGAUAGCCAAAAAUAC ......((((((((((.--.......)))))))...((.((((((....)))))).....(((.((((((.....)))))))))...))..)))........ ( -26.20) >DroEre_CAF1 1674 100 - 1 CAAACAGGUCUCCAGCA--UUACUAAGUUGGAUGAUUCGUGUCCUCUGGAGGACAGUAUGGCUAUGCAAGAUCUGUUUGUUAGCAUAGACAGCCAAAAAUAU ((((((((((((((((.--.......)))))).......((((((....))))))((((....))))..))))))))))....................... ( -28.90) >DroYak_CAF1 1628 100 - 1 CAAACAGGUCUCCAGCA--UAGGAAAGUUGGAUGAUUCGUGUCCUCUGGAGGACAGUAUGGCUAUGCAGGAUCUGUUUGCUAGCAUAGACAGCCAAAAAUAC ...((..(((((((((.--.......)))))).)))..))(((((....)))))....(((((((((((.(......).)).))))....)))))....... ( -29.90) >consensus CAAACAGGUAUCCAGCA__UAACUAAGUUGGAUGAUUCGUGUCCUCUGGAGGACAGUAUGGCUCUGCAAGAUCUGUUUGCUAGCAUAGACAGCCAAAAAUAC (((((((((.((((((..........)))))).......((((((....))))))...............)))))))))....................... (-17.40 = -18.57 + 1.17)
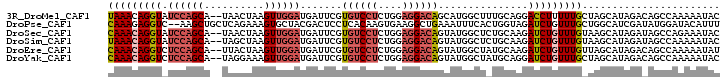
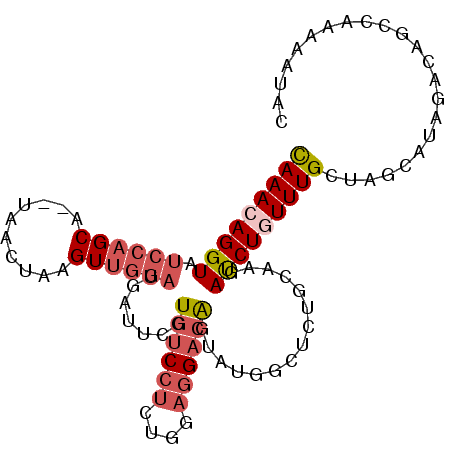
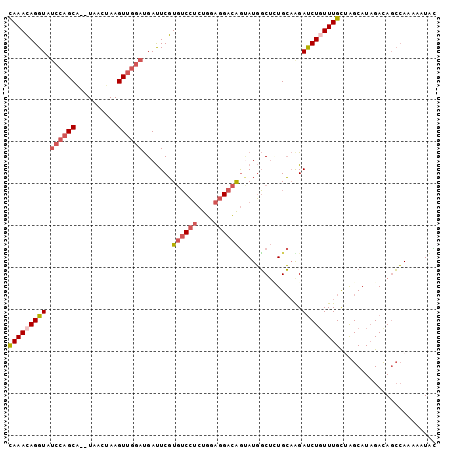
| Location | 20,682,521 – 20,682,619 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 81.08 |
| Mean single sequence MFE | -31.42 |
| Consensus MFE | -17.03 |
| Energy contribution | -18.95 |
| Covariance contribution | 1.92 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.54 |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.639253 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 20682521 98 - 27905053 UCCGAACAUCGAGACGCCAGCUAUGAACAUGAUCUGUAAACAGGUAUCCAGCA--UAACUAAGUUGGAUGAUUCGUGUCCUCUGGAGGACAGCAUGGCUU ((((.....)).)).((((((((((((....(((((....)))))(((((((.--.......)))))))..)))))(((((....)))))))).)))).. ( -32.60) >DroSec_CAF1 1683 98 - 1 UUCGAACAUCGAGACGCCAGCAAUGAAUAUGAUCUGCAAACAGGUAUCCAGCA--UAACUAAGUUGGAUGAUUCGUGUCCUCUGGAGGACAGUAUGGCUC ((((.....))))..((((((.((((((...(((((....)))))(((((((.--.......))))))).))))))(((((....))))).)).)))).. ( -31.60) >DroSim_CAF1 1699 98 - 1 UCCGAACAUCGAGACGCCAGCUAUGAAUAUGAUCUGUAAACAGGUAUCCAGCA--UAGCUAAGUUGGAUGAUUCGUGUCCUCUGGAGGACAGUAUGGCUC ..((((((((..(((...(((((((......(((((....)))))......))--)))))..))).)))).))))((((((....))))))......... ( -33.10) >DroEre_CAF1 1712 98 - 1 UCCGAGUAUCGAGACGCCAGCUAUGAACAUGAUCUGCAAACAGGUCUCCAGCA--UUACUAAGUUGGAUGAUUCGUGUCCUCUGGAGGACAGUAUGGCUA ((((.....)).)).((((((((((((((.((((((....))))))((((((.--.......)))))))).)))))(((((....)))))))).)))).. ( -31.70) >DroYak_CAF1 1666 98 - 1 UCCGUACAUCGAGACGCCAGCUAUGAACCUGAUCUGCAAACAGGUCUCCAGCA--UAGGAAAGUUGGAUGAUUCGUGUCCUCUGGAGGACAGUAUGGCUA .((((((..((((.((((((((........((((((....))))))(((....--..))).)))))).)).))))((((((....))))))))))))... ( -35.60) >DroPer_CAF1 1723 98 - 1 UCCCCAAAUGGAGUCGCCAGCCAUGAAUGAGAUUUGCAAAGAGGUC--AAGCUGCUCAGAAAGUGCUACGACUCCUCACAAGUGAAGGCUGAAAUUUCAC .........(((((((..(((.((...((((....((.........--..))..))))....))))).)))))))......((((((.......)))))) ( -23.90) >consensus UCCGAACAUCGAGACGCCAGCUAUGAACAUGAUCUGCAAACAGGUAUCCAGCA__UAACUAAGUUGGAUGAUUCGUGUCCUCUGGAGGACAGUAUGGCUC ..((.....))....((((((((((((....(((((....))))).((((((..........))))))...)))))(((((....)))))))).)))).. (-17.03 = -18.95 + 1.92)

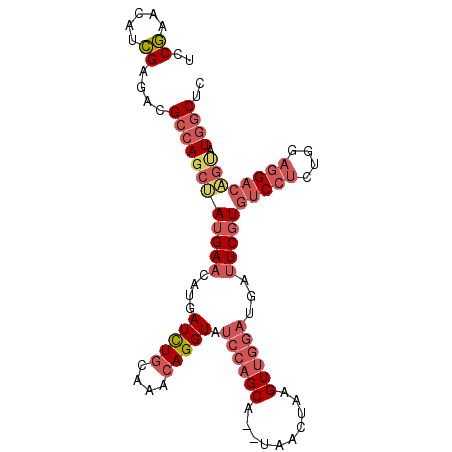
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:52:30 2006