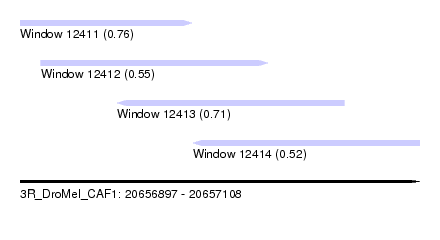
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 20,656,897 – 20,657,108 |
| Length | 211 |
| Max. P | 0.762446 |

| Location | 20,656,897 – 20,656,988 |
|---|---|
| Length | 91 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 83.93 |
| Mean single sequence MFE | -24.32 |
| Consensus MFE | -19.26 |
| Energy contribution | -19.22 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.51 |
| SVM RNA-class probability | 0.762446 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 20656897 91 + 27905053 AUU--------------------------CAUUUUGACCAUACACUAGCCGGCUCACUCUAAUUUCUUGCAAUUAGCAUUACGACUAGUUGGAGUUCGAGAGUUGACCUAAUGUGCU ...--------------------------...........((((.(((.(((((((((((((((((.(((.....)))....))..)))))))))....))))))..))).)))).. ( -19.30) >DroSec_CAF1 13382 91 + 1 AUU--------------------------CAUUUUGAACAUACACGAGCCGGCUCACUCUAAUUUCUUGUAAUUAGCAUUACGACUAGUUGGAGUUCGAGAGUUGACCUAAUGUGCU ...--------------------------...........((((..((.(((((((((((((((..(((((((....)))))))..)))))))))....))))))..))..)))).. ( -22.60) >DroSim_CAF1 4240 91 + 1 AUU--------------------------CAUUUUGAGCAUACACGAGCCGGCUCACUCUAAUUUCUUGUAAUUAGCAUUACGACUAGUUGGAGUUCGAGAGUUGACCUAAUGUGCU ...--------------------------.......((((((....((.(((((((((((((((..(((((((....)))))))..)))))))))....))))))..))..)))))) ( -25.10) >DroEre_CAF1 4177 91 + 1 AUU--------------------------CAUUUUGGAUAUACACGAGUCGGCUCACUCUAAUUUCUUGUAAUUAGCAUUACGGCUAGUUCGAGCUCGAGAGUUGACCUAAUGUGCU ...--------------------------((((..((........(((....)))(((((.....((((.(((((((......)))))))))))....)))))...)).)))).... ( -21.50) >DroYak_CAF1 3714 117 + 1 AUCCAUUGUACAUACAGUGAACGAAGAUCCAUUUUGAAUAUGCACGAGUUGGCUCACUCUAAUUUCUUGUAAUUAGCAUUACAACUAGUUGGAGUUCGAGAGUUGACCUAAUGUGCU ...((((((....))))))......................(((((.((..(((((((((((((..(((((((....)))))))..)))))))))....))))..))....))))). ( -33.10) >consensus AUU__________________________CAUUUUGAACAUACACGAGCCGGCUCACUCUAAUUUCUUGUAAUUAGCAUUACGACUAGUUGGAGUUCGAGAGUUGACCUAAUGUGCU ........................................((((..((.(((((((((((((((..(((((((....)))))))..)))))))))....))))))..))..)))).. (-19.26 = -19.22 + -0.04)

| Location | 20,656,908 – 20,657,028 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.58 |
| Mean single sequence MFE | -32.44 |
| Consensus MFE | -26.44 |
| Energy contribution | -26.44 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.42 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.550799 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 20656908 120 + 27905053 CCAUACACUAGCCGGCUCACUCUAAUUUCUUGCAAUUAGCAUUACGACUAGUUGGAGUUCGAGAGUUGACCUAAUGUGCUCCGCCAUGUAAUUAAUACGUCUGCUGGAGGGAAAAGUAAU ((.((((.(((.(((((((((((((((((.(((.....)))....))..)))))))))....))))))..))).))))((((((.(((((.....)))))..)).))))))......... ( -30.10) >DroSec_CAF1 13393 120 + 1 ACAUACACGAGCCGGCUCACUCUAAUUUCUUGUAAUUAGCAUUACGACUAGUUGGAGUUCGAGAGUUGACCUAAUGUGCUCCGCCAUGUAAUUAAUACGUCUGCUGGAGGGAAAAGUAAU ...((((..((.(((((((((((((((..(((((((....)))))))..)))))))))....))))))..))..))))((((((.(((((.....)))))..)).))))........... ( -31.90) >DroSim_CAF1 4251 120 + 1 GCAUACACGAGCCGGCUCACUCUAAUUUCUUGUAAUUAGCAUUACGACUAGUUGGAGUUCGAGAGUUGACCUAAUGUGCUCCGCCAUGUAAUUAAUACGUCUGCUGGAGGGAAAAGUAAU ...((((..((.(((((((((((((((..(((((((....)))))))..)))))))))....))))))..))..))))((((((.(((((.....)))))..)).))))........... ( -31.90) >DroEre_CAF1 4188 120 + 1 AUAUACACGAGUCGGCUCACUCUAAUUUCUUGUAAUUAGCAUUACGGCUAGUUCGAGCUCGAGAGUUGACCUAAUGUGCUCCGCAGUGUAAUUAAUACGUCUGCUGGAAGGAAAAGUAAU .....((((....((.(((((((.....((((.(((((((......)))))))))))....)))).)))))...)))).(((((((((((.....)))).)))).)))............ ( -31.60) >DroYak_CAF1 3751 120 + 1 AUAUGCACGAGUUGGCUCACUCUAAUUUCUUGUAAUUAGCAUUACAACUAGUUGGAGUUCGAGAGUUGACCUAAUGUGCUCCGCAAUGUAAUUAAUACGUCUGCUGGAGGGAAAAGUAAU ....(((((.((..(((((((((((((..(((((((....)))))))..)))))))))....))))..))....)))))(((((((((((.....))))).))).)))............ ( -36.70) >consensus ACAUACACGAGCCGGCUCACUCUAAUUUCUUGUAAUUAGCAUUACGACUAGUUGGAGUUCGAGAGUUGACCUAAUGUGCUCCGCCAUGUAAUUAAUACGUCUGCUGGAGGGAAAAGUAAU ...((((..((.(((((((((((((((..(((((((....)))))))..)))))))))....))))))..))..))))((((((.(((((.....)))))..)).))))........... (-26.44 = -26.44 + -0.00)

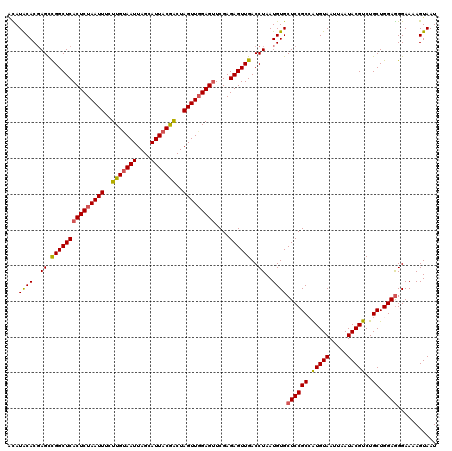
| Location | 20,656,948 – 20,657,068 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.17 |
| Mean single sequence MFE | -28.96 |
| Consensus MFE | -26.16 |
| Energy contribution | -26.20 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.32 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.706883 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 20656948 120 - 27905053 AAAAGGAGGUUAAUUCUAUGCAAUUUUGCAUGGACUCGUGAUUACUUUUCCCUCCAGCAGACGUAUUAAUUACAUGGCGGAGCACAUUAGGUCAACUCUCGAACUCCAACUAGUCGUAAU ....((((......((((((((....)))))))).(((.((((((((....((((.((....(((.....)))...))))))......)))).)).)).))).))))............. ( -28.20) >DroSec_CAF1 13433 120 - 1 AAAAGGAGGUUAAUUCUAUGCAAUUUUGCAUGGACUCGUGAUUACUUUUCCCUCCAGCAGACGUAUUAAUUACAUGGCGGAGCACAUUAGGUCAACUCUCGAACUCCAACUAGUCGUAAU ....((((......((((((((....)))))))).(((.((((((((....((((.((....(((.....)))...))))))......)))).)).)).))).))))............. ( -28.20) >DroSim_CAF1 4291 120 - 1 AAAAGGAGGUUAAUUCUAUGCAAUUUUGCAUGGACUCGUGAUUACUUUUCCCUCCAGCAGACGUAUUAAUUACAUGGCGGAGCACAUUAGGUCAACUCUCGAACUCCAACUAGUCGUAAU ....((((......((((((((....)))))))).(((.((((((((....((((.((....(((.....)))...))))))......)))).)).)).))).))))............. ( -28.20) >DroEre_CAF1 4228 120 - 1 AAAAGGAGGUUAAUUCUAUGCAAUUUUGCAUGGAGUCGUGAUUACUUUUCCUUCCAGCAGACGUAUUAAUUACACUGCGGAGCACAUUAGGUCAACUCUCGAGCUCGAACUAGCCGUAAU .......(((((.(((((((((....))))))((((((.((((((((....((((.((((..(((.....))).))))))))......)))).)).)).))).)))))).)))))..... ( -30.70) >DroYak_CAF1 3791 120 - 1 AAAAGGAGGUUAAUUCUAUGCAAUUUUGCAUGGACUCGUGAUUACUUUUCCCUCCAGCAGACGUAUUAAUUACAUUGCGGAGCACAUUAGGUCAACUCUCGAACUCCAACUAGUUGUAAU ....((((......((((((((....)))))))).(((.((((((((....((((.((((..(((.....))).))))))))......)))).)).)).))).))))............. ( -29.50) >consensus AAAAGGAGGUUAAUUCUAUGCAAUUUUGCAUGGACUCGUGAUUACUUUUCCCUCCAGCAGACGUAUUAAUUACAUGGCGGAGCACAUUAGGUCAACUCUCGAACUCCAACUAGUCGUAAU ....((((......((((((((....)))))))).(((.((((((((....((((.((....(((.....)))...))))))......)))).)).)).))).))))............. (-26.16 = -26.20 + 0.04)

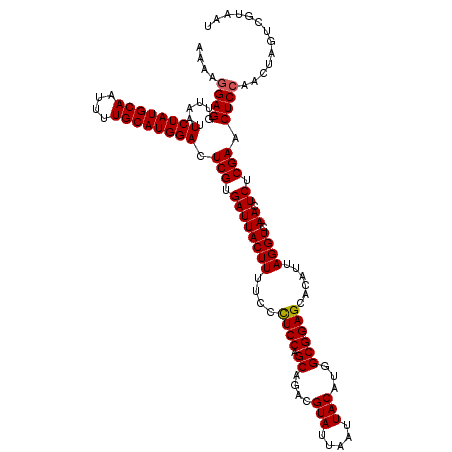
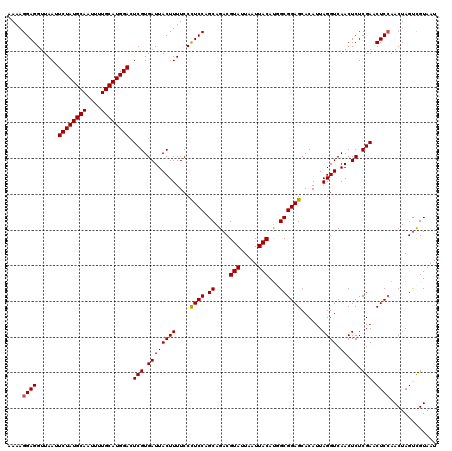
| Location | 20,656,988 – 20,657,108 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.99 |
| Mean single sequence MFE | -32.74 |
| Consensus MFE | -26.00 |
| Energy contribution | -26.84 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.79 |
| SVM decision value | -0.02 |
| SVM RNA-class probability | 0.524466 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 20656988 120 - 27905053 UCGGGGCCCACUUGAACGUGAACGAAAGUCAAUGUACCAUAAAAGGAGGUUAAUUCUAUGCAAUUUUGCAUGGACUCGUGAUUACUUUUCCCUCCAGCAGACGUAUUAAUUACAUGGCGG (((((.....)))))((((..((....))..)))).((((....(((((..((.((((((((....))))))))...((....)).))..))))).......(((.....)))))))... ( -30.80) >DroSec_CAF1 13473 120 - 1 UCGGAGCCCACUUGGACGUGGACGAAAGUCAAUGUACCAUAAAAGGAGGUUAAUUCUAUGCAAUUUUGCAUGGACUCGUGAUUACUUUUCCCUCCAGCAGACGUAUUAAUUACAUGGCGG .....(((....(((((((.(((....))).)))).))).....(((((..((.((((((((....))))))))...((....)).))..))))).......(((.....)))..))).. ( -34.80) >DroSim_CAF1 4331 120 - 1 UCGGAGCCCACUUGGACGUGGACGAAAGUCAAUGUACCAUAAAAGGAGGUUAAUUCUAUGCAAUUUUGCAUGGACUCGUGAUUACUUUUCCCUCCAGCAGACGUAUUAAUUACAUGGCGG .....(((....(((((((.(((....))).)))).))).....(((((..((.((((((((....))))))))...((....)).))..))))).......(((.....)))..))).. ( -34.80) >DroEre_CAF1 4268 119 - 1 UCGAAGC-GACUUAGACGUGGACGAAAGUCAAUGUACCAUAAAAGGAGGUUAAUUCUAUGCAAUUUUGCAUGGAGUCGUGAUUACUUUUCCUUCCAGCAGACGUAUUAAUUACACUGCGG ..((((.-((.....((((.(((....))).)))).......((((..((((((((((((((....))))))))))..))))..))))))))))..((((..(((.....))).)))).. ( -33.00) >DroYak_CAF1 3831 113 - 1 UCGGAGC-GACUU------GGACGAAAGUCAAUGUACCAUAAAAGGAGGUUAAUUCUAUGCAAUUUUGCAUGGACUCGUGAUUACUUUUCCCUCCAGCAGACGUAUUAAUUACAUUGCGG ..((((.-(((..------.(((....)))...))......(((((..((((..((((((((....))))))))....))))..))))).))))).((((..(((.....))).)))).. ( -30.30) >consensus UCGGAGCCCACUUGGACGUGGACGAAAGUCAAUGUACCAUAAAAGGAGGUUAAUUCUAUGCAAUUUUGCAUGGACUCGUGAUUACUUUUCCCUCCAGCAGACGUAUUAAUUACAUGGCGG ...............((((.(((....))).)))).((......(((((..((.((((((((....))))))))...((....)).))..))))).((....(((.....)))...)))) (-26.00 = -26.84 + 0.84)

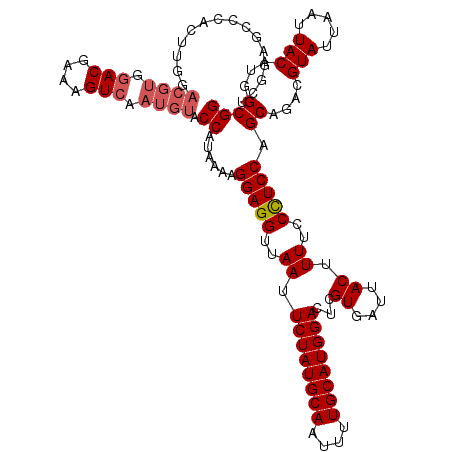
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:52:18 2006