
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 20,641,180 – 20,641,390 |
| Length | 210 |
| Max. P | 0.999068 |

| Location | 20,641,180 – 20,641,270 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 93 |
| Reading direction | reverse |
| Mean pairwise identity | 91.18 |
| Mean single sequence MFE | -22.16 |
| Consensus MFE | -20.06 |
| Energy contribution | -19.90 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.26 |
| Mean z-score | -2.28 |
| Structure conservation index | 0.91 |
| SVM decision value | 3.35 |
| SVM RNA-class probability | 0.999068 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 20641180 90 - 27905053 UUAGGAA---GACGUUCUAGGUCAAGGGACCAAAAGGAAAGAAGACGCUCUAGGCCAAGGGAACAAAAUGAGAGAAGGCGUUCUAGAUCUAGA .......---.....((((((((..(((((.........(((......)))..(((..(....)....(....)..)))))))).)))))))) ( -17.60) >DroSec_CAF1 8996 93 - 1 UCAGAAAGGGGACGUUCUAGGUCAAGGGACCAAAAGGAAAGAAGACGCUCUAGGCCAAGGGAACAAAACGAGAGAAGGCGUUCUAGAUCUAGG ..(((...((((((((((.((((.((((.((....))...(....).)))).))))...)))))....(....).....)))))...)))... ( -21.50) >DroSim_CAF1 9010 93 - 1 UCAGAAAGGGGACGUUCUAGGUCAAGGGACCAAAAGGAAAGAAGACGCUCUAGGCCAAGGGAACAAAACGAGAGAAGGCGUUCUAGAUCUAGG ..(((...((((((((((.((((.((((.((....))...(....).)))).))))...)))))....(....).....)))))...)))... ( -21.50) >DroEre_CAF1 9714 93 - 1 UUAGAAAGAGAACGUUCUAGGUCGAGAGAUCAAAAGGAAAGAAGACGCUCCAGGCCAAGGGAACAAAACGAAAGAAGGCGUUCUAGAUCUAGA (((((...((((((((((.((((((....))....(((..(....)..))).))))...)))))....(....).....)))))...))))). ( -24.60) >DroYak_CAF1 8823 93 - 1 UUAGAAAGAGAACGUUCUAGGUCGAGAGAUCGAAAGGAAAGAAGACGCUCUAGGGCAAGAGAACAAAACGAAAGAAGGCGUUCUAGAUCUAGA (((((...((((((((((.((((....))))...............(((....)))...)))))....(....).....)))))...))))). ( -25.60) >consensus UUAGAAAGAGGACGUUCUAGGUCAAGGGACCAAAAGGAAAGAAGACGCUCUAGGCCAAGGGAACAAAACGAGAGAAGGCGUUCUAGAUCUAGA (((((...((((((((((.((((.((((.((....))...(....).)))).))))...)))))....(....).....)))))...))))). (-20.06 = -19.90 + -0.16)

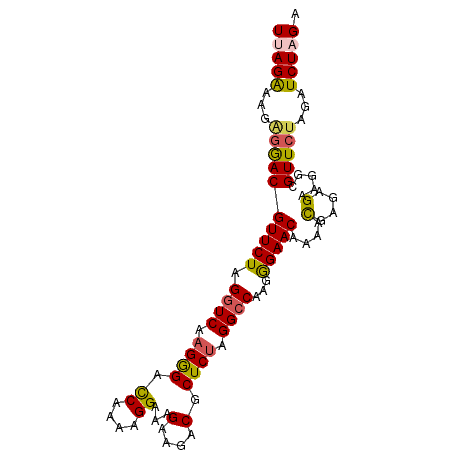

| Location | 20,641,214 – 20,641,310 |
|---|---|
| Length | 96 |
| Sequences | 4 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 69.70 |
| Mean single sequence MFE | -19.91 |
| Consensus MFE | -8.75 |
| Energy contribution | -8.62 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.41 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.44 |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.611641 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 20641214 96 + 27905053 CUUGGCCUAGAGCGUCUUCUUUCCUUUUGGUCCCUUGACCUAGAACGUC---UUCCUAAUUGGUCUCUAGGCCUAGAAGGCCUUCUCUCUUCUCGAUCU ...(((((((((...........((...((((....)))).))......---..((.....)).))))))))).((((((.......))))))...... ( -25.20) >DroEre_CAF1 9748 99 + 1 CUUGGCCUGGAGCGUCUUCUUUCCUUUUGAUCUCUCGACCUAGAACGUUCUCUUUCUAAUUGGUCUCUAGGCCUAGAACGCCUUCUCUCUUCUCGAUCC ((.((((((((..(((............))).....(((((((((.(....).)))))...)))))))))))).))....................... ( -21.70) >DroYak_CAF1 8857 99 + 1 CUUGCCCUAGAGCGUCUUCUUUCCUUUCGAUCUCUCGACCUAGAACGUUCUCUUUCUAAUUGGUCUCUAGGCCUAGAACGCCUUCUCUCUUCUCGAUCC ...((...(((((((..(((......((((....))))...))))))))))..(((((...((((....))))))))).)).................. ( -17.30) >DroAna_CAF1 8221 82 + 1 -----------------UCUUUCGGUUCGAUCCUUUGAUCGGGAACGUCCUUUAUCUGUAUUAUUUUUCGACCAUGACAGUUUUCGGUCAUUGCGACUC -----------------......(((.((......((((((..((((((.....((.............))....))).)))..))))))...))))). ( -15.42) >consensus CUUGGCCUAGAGCGUCUUCUUUCCUUUCGAUCCCUCGACCUAGAACGUCCUCUUUCUAAUUGGUCUCUAGGCCUAGAACGCCUUCUCUCUUCUCGAUCC ............................((((....(((((((((........)))))...))))...((((.......))))...........)))). ( -8.75 = -8.62 + -0.12)

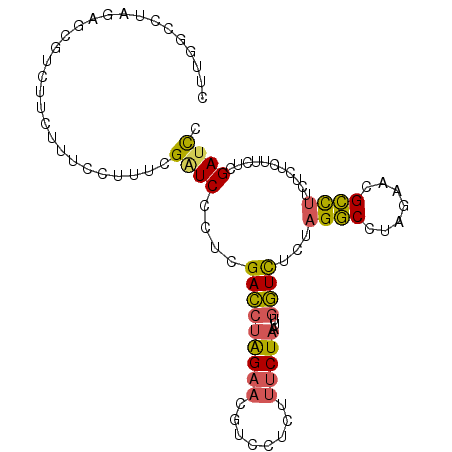
| Location | 20,641,214 – 20,641,310 |
|---|---|
| Length | 96 |
| Sequences | 4 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 69.70 |
| Mean single sequence MFE | -22.05 |
| Consensus MFE | -14.29 |
| Energy contribution | -16.48 |
| Covariance contribution | 2.19 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.65 |
| SVM decision value | 1.59 |
| SVM RNA-class probability | 0.966118 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 20641214 96 - 27905053 AGAUCGAGAAGAGAGAAGGCCUUCUAGGCCUAGAGACCAAUUAGGAA---GACGUUCUAGGUCAAGGGACCAAAAGGAAAGAAGACGCUCUAGGCCAAG ......(((((.(......)))))).(((((((((......(((((.---....)))))((((....))))................)))))))))... ( -29.10) >DroEre_CAF1 9748 99 - 1 GGAUCGAGAAGAGAGAAGGCGUUCUAGGCCUAGAGACCAAUUAGAAAGAGAACGUUCUAGGUCGAGAGAUCAAAAGGAAAGAAGACGCUCCAGGCCAAG .................(((.((((.(((((((((.....((.....)).....))))))))).)))).......(((..(....)..)))..)))... ( -22.70) >DroYak_CAF1 8857 99 - 1 GGAUCGAGAAGAGAGAAGGCGUUCUAGGCCUAGAGACCAAUUAGAAAGAGAACGUUCUAGGUCGAGAGAUCGAAAGGAAAGAAGACGCUCUAGGGCAAG (((.((.............)).))).(.(((((((......(((((........)))))((((....))))................))))))).)... ( -22.32) >DroAna_CAF1 8221 82 - 1 GAGUCGCAAUGACCGAAAACUGUCAUGGUCGAAAAAUAAUACAGAUAAAGGACGUUCCCGAUCAAAGGAUCGAACCGAAAGA----------------- ...((((.(((((........))))).).))).................((.....))(((((....)))))...(....).----------------- ( -14.10) >consensus GGAUCGAGAAGAGAGAAGGCGUUCUAGGCCUAGAGACCAAUUAGAAAGAGAACGUUCUAGGUCAAGAGAUCAAAAGGAAAGAAGACGCUCUAGGCCAAG .........((((........)))).(((((((((......(((((........)))))((((....))))................)))))))))... (-14.29 = -16.48 + 2.19)

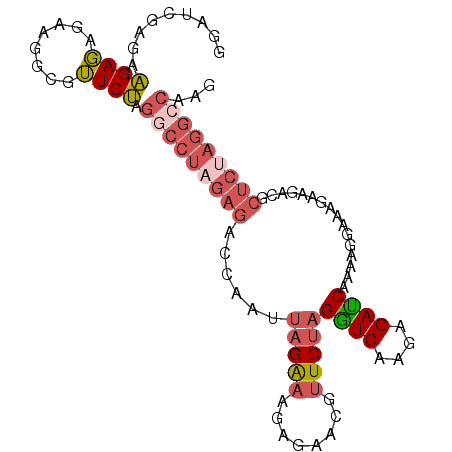
| Location | 20,641,243 – 20,641,350 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 90.55 |
| Mean single sequence MFE | -31.02 |
| Consensus MFE | -25.10 |
| Energy contribution | -25.66 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.81 |
| SVM decision value | 1.51 |
| SVM RNA-class probability | 0.959980 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 20641243 107 - 27905053 GGUCUCGGGAUGCAACACAUAGAAGGUACUCUAGGUCAAGAGAUCGAGAAGAGAGAAGGCCUUCUAGGCCUAGAGACCAAUUAGGAA---GACGUUCUAGGUCAAGGGAC .(((((.............((((((((.((((.((((....))))......))))...))))))))(((((((((.((.....))..---....)))))))))..))))) ( -35.60) >DroSec_CAF1 9059 110 - 1 GGUCUCGGGAUACAACACAAAGAAGGCAUUCUAGGUCAAGAGAUCGAGAAGACAGAAGGCGUUCUAGGCCUAGAGACCAAUCAGAAAGGGGACGUUCUAGGUCAAGGGAC .(((((..............((((.((.((((.((((....)))).))))........)).)))).(((((((((.((..........))....)))))))))..))))) ( -31.50) >DroSim_CAF1 9073 110 - 1 GGUCUCGGGAUACAACACAAAGAAGGCACUCUAGGUCAAGAGAUCGAGAAGACAGAAGGCGUUCUAGGCCUAGAGACCAAUCAGAAAGGGGACGUUCUAGGUCAAGGGAC .(((((..(((.............((..(((((((((...(((.((.............)).))).))))))))).))....((((.(....).))))..)))..))))) ( -31.82) >DroEre_CAF1 9777 110 - 1 GGUCACGGGAUAGAACACAUAGAAGACAUUCUAGGUCAAGGGAUCGAGAAGAGAGAAGGCGUUCUAGGCCUAGAGACCAAUUAGAAAGAGAACGUUCUAGGUCGAGAGAU ((((.((((.((((((.(.(((((....)))))((((....)))).............).))))))..))).).))))..((((((........))))))(((....))) ( -28.10) >DroYak_CAF1 8886 110 - 1 GGUCUCGGGAUACAACACACAGAAGGCAUUCUAAGUCAAGGGAUCGAGAAGAGAGAAGGCGUUCUAGGCCUAGAGACCAAUUAGAAAGAGAACGUUCUAGGUCGAGAGAU .(((((..............((((.((.((((...((..(....)..))....)))).)).)))).(((((((((.....((.....)).....)))))))))..))))) ( -28.10) >consensus GGUCUCGGGAUACAACACAAAGAAGGCAUUCUAGGUCAAGAGAUCGAGAAGAGAGAAGGCGUUCUAGGCCUAGAGACCAAUUAGAAAGAGGACGUUCUAGGUCAAGGGAC .(((((..............((((.((.((((.((((....)))).))))........)).)))).(((((((((.....((.....)).....)))))))))..))))) (-25.10 = -25.66 + 0.56)


| Location | 20,641,270 – 20,641,390 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.56 |
| Mean single sequence MFE | -30.64 |
| Consensus MFE | -22.54 |
| Energy contribution | -23.98 |
| Covariance contribution | 1.44 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.74 |
| SVM decision value | -0.04 |
| SVM RNA-class probability | 0.515135 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 20641270 120 - 27905053 UCAAAUCGCGAAAGGAAUCGCAAUGAAAAAAGGUCAUCGAGGUCUCGGGAUGCAACACAUAGAAGGUACUCUAGGUCAAGAGAUCGAGAAGAGAGAAGGCCUUCUAGGCCUAGAGACCAA .......((((......))))..........((((.((.(((((..(........)...((((((((.((((.((((....))))......))))...))))))))))))).)))))).. ( -37.00) >DroSec_CAF1 9089 120 - 1 UCAAAUCGCGAAAGGAAUCGCACUGAAAAAAGGUCAUCGAGGUCUCGGGAUACAACACAAAGAAGGCAUUCUAGGUCAAGAGAUCGAGAAGACAGAAGGCGUUCUAGGCCUAGAGACCAA .......((((......))))..........((((.((.((((((.(.....).......((((.((.((((.((((....)))).))))........)).)))))))))).)))))).. ( -31.90) >DroSim_CAF1 9103 120 - 1 UCAAAUCGCGAAAGGAAUCGCUAUGAAAAAAGGUCAUCGAGGUCUCGGGAUACAACACAAAGAAGGCACUCUAGGUCAAGAGAUCGAGAAGACAGAAGGCGUUCUAGGCCUAGAGACCAA .......((((......))))..........((((.((.((((((.(.....).......((((.((..(((.((((....))))(......))))..)).)))))))))).)))))).. ( -30.10) >DroEre_CAF1 9807 117 - 1 UCUGAUCGGGAAAGGAUUCGCAAUGAG---AGAUCAUCAAGGUCACGGGAUAGAACACAUAGAAGACAUUCUAGGUCAAGGGAUCGAGAAGAGAGAAGGCGUUCUAGGCCUAGAGACCAA (((.((.(.(((....))).).)).))---).........((((.((((.((((((.(.(((((....)))))((((....)))).............).))))))..))).).)))).. ( -28.30) >DroYak_CAF1 8916 117 - 1 UCGGAUCGGGAAAGGAAUCGCAACGAG---AGAUCAUCAAGGUCUCGGGAUACAACACACAGAAGGCAUUCUAAGUCAAGGGAUCGAGAAGAGAGAAGGCGUUCUAGGCCUAGAGACCAA ..((.(((((...((((.(((.....(---(((((.....))))))......................((((..(((....)))..))))........)))))))...))).))..)).. ( -25.90) >consensus UCAAAUCGCGAAAGGAAUCGCAAUGAAAAAAGGUCAUCGAGGUCUCGGGAUACAACACAAAGAAGGCAUUCUAGGUCAAGAGAUCGAGAAGAGAGAAGGCGUUCUAGGCCUAGAGACCAA .......((((......))))..........((((.((.(((((..(........)....((((.((.((((.((((....)))).))))........)).)))).))))).)))))).. (-22.54 = -23.98 + 1.44)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:52:07 2006