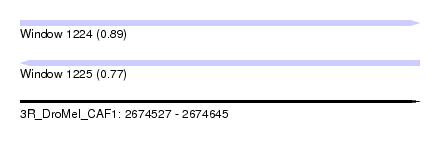
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 2,674,527 – 2,674,645 |
| Length | 118 |
| Max. P | 0.886619 |

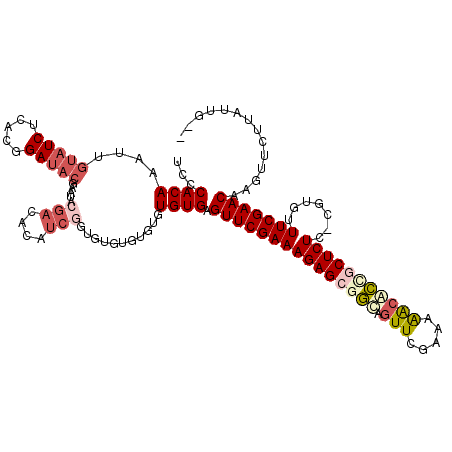
| Location | 2,674,527 – 2,674,645 |
|---|---|
| Length | 118 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 77.97 |
| Mean single sequence MFE | -35.34 |
| Consensus MFE | -19.71 |
| Energy contribution | -21.29 |
| Covariance contribution | 1.58 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.13 |
| Structure conservation index | 0.56 |
| SVM decision value | 0.94 |
| SVM RNA-class probability | 0.886619 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 2674527 118 + 27905053 UCCCACAAAUUGUAUCUCACGGAUACGAAUACGACACAUCGGUGUGUGUGUGUGUGAGUUCGAAAGAGCGGUAGUUCGAAAAGCACCGCUCUCGCGUGUUUCGAACAAGUUCUUAUUG-- ...((((..(((((((.....)))))))((((.(((((....))))))))).)))).(((((((((((((((.(((.....))))))))))).......)))))))............-- ( -38.61) >DroSec_CAF1 33960 92 + 1 UCCCACAAAUUGUA------------------------UCGCU--GUGUGUGUGUGAGUUCGAAAGAGCAGCAGUUCGAAAAACACCUCUCUCGCGUGUUUCGAACAAGUUCUUAUUG-- ...((((....(..------------------------...).--...))))(((..((((....)))).)))(((((((..(((((......).)))))))))))............-- ( -22.40) >DroSim_CAF1 27061 116 + 1 UCCCACAAAUUGUAUCUCACGGAUACGAAUACGACACAUCGCU--GUGUGUGUGUGAGUUCGAAAGAGCAGCAGUUCGAAAAACACCGCUCUCGCGUGUUUCGAACAAGUUCUUAUUG-- ...(((...(((((((.....)))))))((((.(((((....)--))))))))))).((((....))))....(((((((..((((.((....)))))))))))))............-- ( -36.60) >DroEre_CAF1 27214 115 + 1 UGCCACAAAGUGUAUCUCACGGAUACGUAUCCGAGACAUCGGUGUGUG--CGUGUGAGUUCGAAAGAGCGGUUGUUCGAAAAGCACCGCUCUC-CGCUUUUCGAACAAGUUCUUAUUG-- .(((..........((.((((.(((((...((((....))))))))).--)))).))((((....)))))))((((((((((((.........-.))))))))))))...........-- ( -43.00) >DroYak_CAF1 34361 117 + 1 UGCCACAAAGUGUAUCUCACGGAUACGUAUACGACACAUCGGUGUGUGUGUGUGUGAGUUCGAAAGAGCGGUUGUUAGAAAAGCACCGCUCUC-CGCUUUUCGAACAAGUUCUUAUUG-- ...((((....(((((.....))))).(((((.(((((....)))))))))))))).(((((((((((((((.(((.....))))))))))).-.....)))))))............-- ( -38.30) >DroAna_CAF1 22049 105 + 1 AGCCACAAAUUGUAUCUCACGGAUACCCA-----------GGCCCGUGUGCGUGUGAGUUCGAAAGAGUGCCACUUCGAAAGAUGGUCCUCUC----AUUUCGAACAAGUUCUUAUUGCU (((...........((.((((.((((...-----------.....)))).)))).))(((((((((((..(((..((....)))))..)))).----..)))))))...........))) ( -33.10) >consensus UCCCACAAAUUGUAUCUCACGGAUACGAAUACGACACAUCGGUGUGUGUGUGUGUGAGUUCGAAAGAGCGGCAGUUCGAAAAACACCGCUCUC_CGUGUUUCGAACAAGUUCUUAUUG__ ...((((....(((((.....))))).....(((....)))...........)))).(((((((((((((((.(((.....))))))))))).......))))))).............. (-19.71 = -21.29 + 1.58)

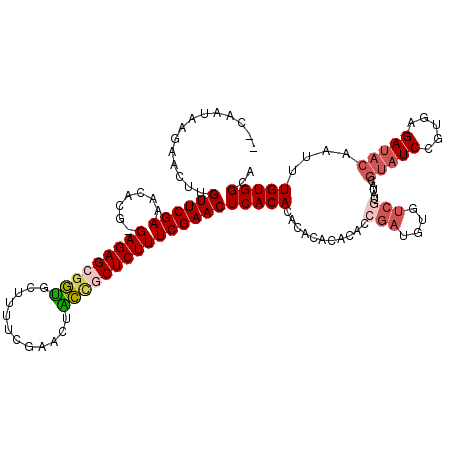
| Location | 2,674,527 – 2,674,645 |
|---|---|
| Length | 118 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.97 |
| Mean single sequence MFE | -31.04 |
| Consensus MFE | -20.51 |
| Energy contribution | -21.51 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.52 |
| SVM RNA-class probability | 0.766223 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 2674527 118 - 27905053 --CAAUAAGAACUUGUUCGAAACACGCGAGAGCGGUGCUUUUCGAACUACCGCUCUUUCGAACUCACACACACACACACCGAUGUGUCGUAUUCGUAUCCGUGAGAUACAAUUUGUGGGA --......(((....)))......((.((((((((((.((....)).)))))))))).))..((((((.(((((((......))))).))....((((.(....)))))....)))))). ( -34.60) >DroSec_CAF1 33960 92 - 1 --CAAUAAGAACUUGUUCGAAACACGCGAGAGAGGUGUUUUUCGAACUGCUGCUCUUUCGAACUCACACACACAC--AGCGA------------------------UACAAUUUGUGGGA --....((((.(..((((((((.((((.......)))).))))))))....).)))).....((((((.......--.....------------------------.......)))))). ( -19.31) >DroSim_CAF1 27061 116 - 1 --CAAUAAGAACUUGUUCGAAACACGCGAGAGCGGUGUUUUUCGAACUGCUGCUCUUUCGAACUCACACACACAC--AGCGAUGUGUCGUAUUCGUAUCCGUGAGAUACAAUUUGUGGGA --......(((....)))......((.(((((((((((((...)))).))))))))).))..((((((.((((((--(....))))).))....((((.(....)))))....)))))). ( -34.20) >DroEre_CAF1 27214 115 - 1 --CAAUAAGAACUUGUUCGAAAAGCG-GAGAGCGGUGCUUUUCGAACAACCGCUCUUUCGAACUCACACG--CACACACCGAUGUCUCGGAUACGUAUCCGUGAGAUACACUUUGUGGCA --............((((((......-(((((((((............)))))))))))))))......(--(.((((((((....))))....((((.(....)))))....)))))). ( -36.10) >DroYak_CAF1 34361 117 - 1 --CAAUAAGAACUUGUUCGAAAAGCG-GAGAGCGGUGCUUUUCUAACAACCGCUCUUUCGAACUCACACACACACACACCGAUGUGUCGUAUACGUAUCCGUGAGAUACACUUUGUGGCA --............((((((......-(((((((((............)))))))))))))))...((((.(((((((....))))).))....((((.(....)))))....))))... ( -32.70) >DroAna_CAF1 22049 105 - 1 AGCAAUAAGAACUUGUUCGAAAU----GAGAGGACCAUCUUUCGAAGUGGCACUCUUUCGAACUCACACGCACACGGGCC-----------UGGGUAUCCGUGAGAUACAAUUUGUGGCU (((.(((((.....((((((...----(((((..((((........))))..))))))))))).........(((((((.-----------...))..)))))........))))).))) ( -29.30) >consensus __CAAUAAGAACUUGUUCGAAACACG_GAGAGCGGUGCUUUUCGAACUACCGCUCUUUCGAACUCACACACACACACACCGAUGUGUCGUAUACGUAUCCGUGAGAUACAAUUUGUGGCA ..............((((((.......(((((((((............)))))))))))))))(((((...........(((....))).....(((((.....)))))....))))).. (-20.51 = -21.51 + 1.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:51:38 2006