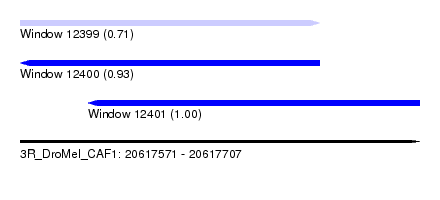
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 20,617,571 – 20,617,707 |
| Length | 136 |
| Max. P | 0.997043 |

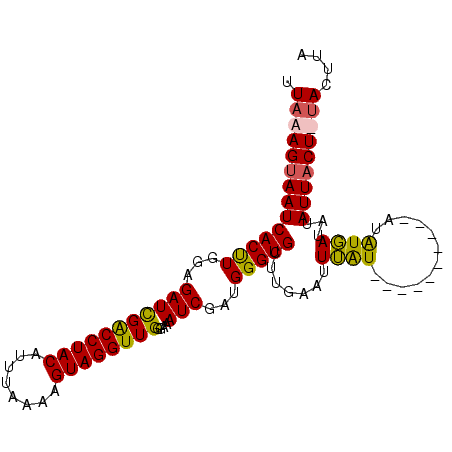
| Location | 20,617,571 – 20,617,673 |
|---|---|
| Length | 102 |
| Sequences | 4 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 81.71 |
| Mean single sequence MFE | -22.45 |
| Consensus MFE | -16.40 |
| Energy contribution | -16.02 |
| Covariance contribution | -0.37 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.706987 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 20617571 102 + 27905053 CUAAAGCAAUCACUUGAAGAUCGACCUACAUUCAAAAGUAGGUUGGGAAAUCGAUGGGUGUUUGAAUUUGUUUGGAUUUCAAUAUGAUAUAUUACU-UAGUUU ........((((........(((((((((........)))))))))(((((((((..((......))..)))..))))))....))))........-...... ( -21.40) >DroSec_CAF1 3549 93 + 1 UUAAAGUAAUCACUUGGAGAUCGACCUACAUUUAAAAGUAGGUUGGGAAAUCGAAGGGUGCUUGAAUUUAU----------AUAUGAUAUAUUACUUUACUUA .((((((((((.(((.((..(((((((((........)))))))))....)).)))))).........(((----------((...))))).))))))).... ( -22.00) >DroSim_CAF1 3528 93 + 1 UUAAAGUAAUCACUUGGAGAUCGACCUACAUUUAAAAGUAGGUUGGGAAAUCGAUGGGUGCUUGAAUUUAU----------AUAUGAUAUAUUACUUUACUUA .((((((((.((((((..(((((((((((........))))))))....)))..))))))........(((----------((...))))))))))))).... ( -20.90) >DroYak_CAF1 3820 102 + 1 UUAAAGUAAUCACCUGGCGAUUGGCCUACAUUGCAAAGUAGGUUGGGAAAUCGAUGGGUGUUGGAAUUGGUUUGGAUUUUAAUGUCAUAUAUUACU-CAGGUU ...........((((((((((..((((((........))))))..)....)))...((..((((((((......))))))))..)).........)-))))). ( -25.50) >consensus UUAAAGUAAUCACUUGGAGAUCGACCUACAUUUAAAAGUAGGUUGGGAAAUCGAUGGGUGCUUGAAUUUAU__________AUAUGAUAUAUUACU_UACUUA .((((((((((((((...(((((((((((........))))))))....)))...))))).......((((............))))...))))))))).... (-16.40 = -16.02 + -0.37)

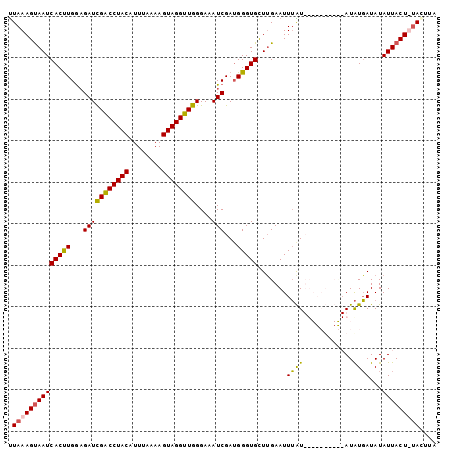
| Location | 20,617,571 – 20,617,673 |
|---|---|
| Length | 102 |
| Sequences | 4 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 81.71 |
| Mean single sequence MFE | -15.76 |
| Consensus MFE | -11.41 |
| Energy contribution | -12.47 |
| Covariance contribution | 1.06 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.14 |
| Structure conservation index | 0.72 |
| SVM decision value | 1.22 |
| SVM RNA-class probability | 0.932465 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 20617571 102 - 27905053 AAACUA-AGUAAUAUAUCAUAUUGAAAUCCAAACAAAUUCAAACACCCAUCGAUUUCCCAACCUACUUUUGAAUGUAGGUCGAUCUUCAAGUGAUUGCUUUAG ...(((-((((((........(((((...........))))).(((.....((((.....((((((........)))))).)))).....)))))))).)))) ( -15.50) >DroSec_CAF1 3549 93 - 1 UAAGUAAAGUAAUAUAUCAUAU----------AUAAAUUCAAGCACCCUUCGAUUUCCCAACCUACUUUUAAAUGUAGGUCGAUCUCCAAGUGAUUACUUUAA ....(((((((....(((((..----------..(((((.(((....))).)))))..(.((((((........)))))).)........)))))))))))). ( -15.70) >DroSim_CAF1 3528 93 - 1 UAAGUAAAGUAAUAUAUCAUAU----------AUAAAUUCAAGCACCCAUCGAUUUCCCAACCUACUUUUAAAUGUAGGUCGAUCUCCAAGUGAUUACUUUAA ....(((((((((((((...))----------)))...(((.......(((((........(((((........)))))))))).......))))))))))). ( -15.14) >DroYak_CAF1 3820 102 - 1 AACCUG-AGUAAUAUAUGACAUUAAAAUCCAAACCAAUUCCAACACCCAUCGAUUUCCCAACCUACUUUGCAAUGUAGGCCAAUCGCCAGGUGAUUACUUUAA .....(-((((((....((........))..............((((...(((((......(((((........)))))..)))))...)))))))))))... ( -16.70) >consensus AAACUA_AGUAAUAUAUCAUAU__________ACAAAUUCAAACACCCAUCGAUUUCCCAACCUACUUUUAAAUGUAGGUCGAUCUCCAAGUGAUUACUUUAA ....(((((((((......((((........))))........(((.....((((.....((((((........)))))).)))).....)))))))))))). (-11.41 = -12.47 + 1.06)

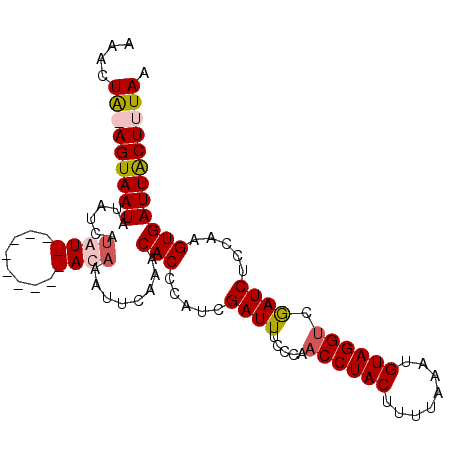

| Location | 20,617,594 – 20,617,707 |
|---|---|
| Length | 113 |
| Sequences | 4 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 76.28 |
| Mean single sequence MFE | -17.88 |
| Consensus MFE | -10.24 |
| Energy contribution | -12.55 |
| Covariance contribution | 2.31 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.83 |
| Structure conservation index | 0.57 |
| SVM decision value | 2.79 |
| SVM RNA-class probability | 0.997043 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 20617594 113 - 27905053 CAAUGGAA--UGUCUAUCUUACUUAACUUGCACUAGAAACUA-AGUAAUAUAUCAUAUUGAAAUCCAAACAAAUUCAAACACCCAUCGAUUUCCCAACCUACUUUUGAAUGUAGGU ...(((((--(((.(((.(((((((...............))-))))).)))..)))))((((((......................)))))))))((((((........)))))) ( -16.71) >DroSec_CAF1 3572 106 - 1 CGAUAGACAUUCCCUAUCUUACUUAACUUGCACUAGUAAGUAAAGUAAUAUAUCAUAU----------AUAAAUUCAAGCACCCUUCGAUUUCCCAACCUACUUUUAAAUGUAGGU .(((((.......)))))((((((.((((((....)))))).))))))..........----------..(((((.(((....))).)))))....((((((........)))))) ( -22.20) >DroSim_CAF1 3551 106 - 1 CGAUAGACAUUCCCUAUCUUACUUAACUUGCACUAGUAAGUAAAGUAAUAUAUCAUAU----------AUAAAUUCAAGCACCCAUCGAUUUCCCAACCUACUUUUAAAUGUAGGU .(((((.......)))))((((((.((((((....)))))).))))))..........----------............................((((((........)))))) ( -21.80) >DroYak_CAF1 3843 105 - 1 CCCUU----------AUCGAACUUAUCUUUUACAAGAACCUG-AGUAAUAUAUGACAUUAAAAUCCAAACCAAUUCCAACACCCAUCGAUUUCCCAACCUACUUUGCAAUGUAGGC .....----------((((((((((((((....))))...))-))).......((........))....................))))).......(((((........))))). ( -10.80) >consensus CGAUAGAC__UCCCUAUCUUACUUAACUUGCACUAGAAACUA_AGUAAUAUAUCAUAU__________ACAAAUUCAAACACCCAUCGAUUUCCCAACCUACUUUUAAAUGUAGGU ..............(((.((((((.((((((....)))))).)))))).)))............................................((((((........)))))) (-10.24 = -12.55 + 2.31)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:52:01 2006