
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 20,616,865 – 20,617,009 |
| Length | 144 |
| Max. P | 0.764646 |

| Location | 20,616,865 – 20,616,969 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 84.52 |
| Mean single sequence MFE | -29.18 |
| Consensus MFE | -23.40 |
| Energy contribution | -23.90 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.29 |
| SVM RNA-class probability | 0.670495 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
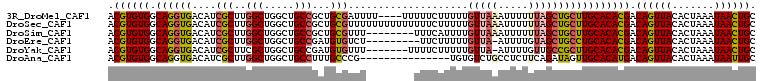
>3R_DroMel_CAF1 20616865 104 + 27905053 ACGUGUGGCAGGUGACAUCGCUUGGCUGGCUGCCGCUGCGAUUUU----UUUUUCUUUUUGUUAAAUUUUUUAGCUGCUUGCACACGACAGUUACACUAAAUAACUGC .((((((.(((((...(((((.((((.....))))..)))))...----...........(((((.....))))).))))))))))).((((((.......)))))). ( -31.80) >DroSec_CAF1 2839 108 + 1 ACGUGUGGCAGGUGACAUCGCUUGGCUGGCUGCCGCUGCGUUUUUUUUUUUUUUCUUUUUGUUAAAUUUUUUAGCUGCUUGCACACGACAGUUACACUAAAUAACUGC .((((((.(((((.....(((.((((.....))))..)))....................(((((.....))))).))))))))))).((((((.......)))))). ( -29.10) >DroSim_CAF1 2826 100 + 1 ACGUGUGGCAGGUGACAUCGCUUGGCUGGCUGCCGCUGCGUUU--------UUUCAUUUUGUUAAAUUUUUUAGCUGCUUGCACACGACAGUUACACUAAAUAACUGC .((((((.(((((((...(((.((((.....))))..)))...--------..)).....(((((.....))))).))))))))))).((((((.......)))))). ( -29.20) >DroEre_CAF1 3341 98 + 1 ACGUGUGGCAGGUGACAUCGCUUGGCUGGCUGCCGAUGUGUCU---------UUCUUUUUGUUA-AUUUUGUAGCUGCCUGCACACGACAGUUACACUAAAUAACUGC .((((((.(((((((((.((.(((((.....))))))))))).---------........((((-......)))).))))))))))).((((((.......)))))). ( -33.30) >DroYak_CAF1 3141 100 + 1 ACGUGUGGCAGGUGACAUCGCUUCGCUGGCUGCCGAUGUGUUU-------UUUUCUUUUUGUUA-AUUUUGUUGCCGCUUGCACACGACAGUUACACUAAAUAACUGC .((((((.((((((((((((....((.....))))))))....-------..........((..-........)))))))))))))).((((((.......)))))). ( -26.00) >DroAna_CAF1 2583 93 + 1 ACGUGUGGCAGGUGACAUCGCUUGGCUGGCUGCCUUUGCCCG---------------UGUGUCUGCCUCUUCAGAUAGUUGCACAUGACAGUUACACUAAAUAAUUGC ..(((((((.(....)...((..(((.....)))...)).((---------------((((((((.((....)).)))..)))))))...)))))))........... ( -25.70) >consensus ACGUGUGGCAGGUGACAUCGCUUGGCUGGCUGCCGCUGCGUUU________UUUCUUUUUGUUAAAUUUUUUAGCUGCUUGCACACGACAGUUACACUAAAUAACUGC .((((((.((((((....(((..(((.....)))...)))....................(((((.....))))))))))))))))).((((((.......)))))). (-23.40 = -23.90 + 0.50)
| Location | 20,616,902 – 20,617,009 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 78.31 |
| Mean single sequence MFE | -20.28 |
| Consensus MFE | -13.43 |
| Energy contribution | -14.15 |
| Covariance contribution | 0.72 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.51 |
| SVM RNA-class probability | 0.764646 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
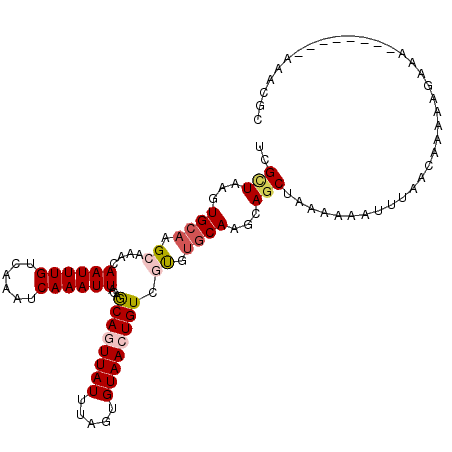
>3R_DroMel_CAF1 20616902 107 - 27905053 UCGUUAAGUGCAAGCAAACAAUUUGUCAAAUCAAAUUACAGCAGUUAUUUAGUGUAACUGUCGUGUGCAAGCAGCUAAAAAAUUUAACAAAAAGAAAAA----AAAAUCGC ..(((((((...(((....((((((......))))))((.((((((((.....)))))))).)).........))).....)))))))...........----........ ( -20.20) >DroSec_CAF1 2876 111 - 1 UCGUUAAGUGCAAGCAAACAAUUUGUCAAAUCAAAUUACAGCAGUUAUUUAGUGUAACUGUCGUGUGCAAGCAGCUAAAAAAUUUAACAAAAAGAAAAAAAAAAAAAACGC ..(((((((...(((....((((((......))))))((.((((((((.....)))))))).)).........))).....)))))))....................... ( -20.20) >DroSim_CAF1 2863 103 - 1 UCGUUAAGUGCAAGCAAACAAUUUGUCAAAUCAAAUUACAGCAGUUAUUUAGUGUAACUGUCGUGUGCAAGCAGCUAAAAAAUUUAACAAAAUGAAA--------AAACGC ..(((((((...(((....((((((......))))))((.((((((((.....)))))))).)).........))).....))))))).........--------...... ( -20.20) >DroEre_CAF1 3378 101 - 1 UCGCUAAGUGCAAGCAAACAAUUUGUCAAAUCAAAUUACGGCAGUUAUUUAGUGUAACUGUCGUGUGCAGGCAGCUACAAAAU-UAACAAAAAGAA---------AGACAC ......(((((..(((...((((((......))))))(((((((((((.....))))))))))).)))..))).)).......-............---------...... ( -24.30) >DroAna_CAF1 2620 96 - 1 UUGCUAAGUGCAAGCAAACAAUUUGUCAAAUCAAAUUACCGCAAUUAUUUAGUGUAACUGUCAUGUGCAACUAUCUGAAGAGGCAGACACA---------------CGGGC ((((.....))))((....((((((......))))))...)).........((((..(((((.....((......))....)))))..)))---------------).... ( -16.90) >DroPer_CAF1 2885 105 - 1 UUGCUAAGUGCAAGCAAACAAUUUGUCAAAUCAAAUUACCACAAUUAUUUAGUGUAACUGUCACGAUCACAUAGCGCAC---UCGGAGGCAGUGGCA---GAGGCAGAGGC (((((.......)))))....((((((............(((.........)))...(((((((..((.(..((....)---)..).))..))))))---).))))))... ( -19.90) >consensus UCGCUAAGUGCAAGCAAACAAUUUGUCAAAUCAAAUUACAGCAGUUAUUUAGUGUAACUGUCGUGUGCAAGCAGCUAAAAAAUUUAACAAAAAGAAA________AAACGC ..(((...((((.((....((((((......))))))...((((((((.....)))))))).)).))))...))).................................... (-13.43 = -14.15 + 0.72)

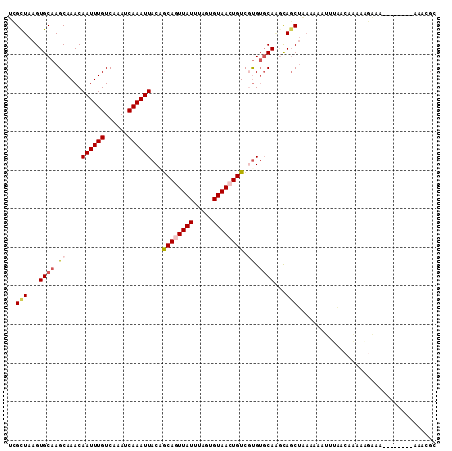
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:51:59 2006