
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 20,594,761 – 20,594,892 |
| Length | 131 |
| Max. P | 0.855134 |

| Location | 20,594,761 – 20,594,854 |
|---|---|
| Length | 93 |
| Sequences | 3 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 85.15 |
| Mean single sequence MFE | -12.95 |
| Consensus MFE | -9.32 |
| Energy contribution | -9.10 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.570866 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 20594761 93 + 27905053 UAACGACAUCCGCACACUUUACAUAU--AUAUUUGGAACCU----CCAAUGGAUUCUGU-UCCUAAAAAUCCACACACAUAUGCUGGAAUUUUAUCU---UUU ........((((((............--....((((.....----))))((((((.(..-.....).))))))........))).))).........---... ( -13.10) >DroSec_CAF1 19564 98 + 1 UAACGACAUCAGCACACCUUACAUAU--AUAUUCGGAACCUCCAUCCAAUGGAUCCCGUUUACUAAAAAUCCACACUAAUAUGCUGGACUUUUAUCU---UUU ........((((((............--......(((.......)))..(((((..............)))))........))))))..........---... ( -12.14) >DroSim_CAF1 19678 102 + 1 UAACAACAUCAGUACAUUUUACAUAUAUAUAUUUGGAACCUCCCUCCAAUGGAUCCUGU-UACUAAAAAUCCACACACAUAUGCUGGACUUUUAUCUCUCUUU (((((..(((.(((.....)))..........(((((.......)))))..)))..)))-)).(((((.((((((......)).)))).)))))......... ( -13.60) >consensus UAACGACAUCAGCACACUUUACAUAU__AUAUUUGGAACCUCC_UCCAAUGGAUCCUGU_UACUAAAAAUCCACACACAUAUGCUGGACUUUUAUCU___UUU ..................................(((.((..........)).))).......(((((.((((((......)).)))).)))))......... ( -9.32 = -9.10 + -0.22)

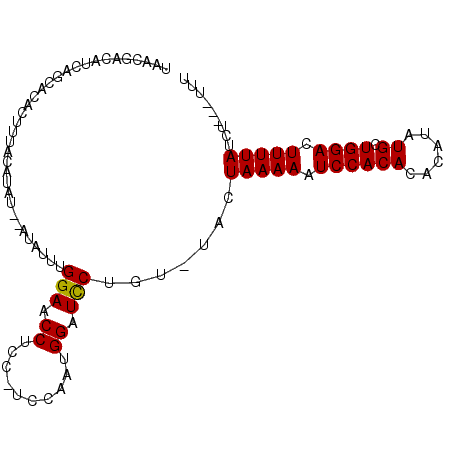

| Location | 20,594,761 – 20,594,854 |
|---|---|
| Length | 93 |
| Sequences | 3 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 85.15 |
| Mean single sequence MFE | -23.20 |
| Consensus MFE | -16.23 |
| Energy contribution | -16.13 |
| Covariance contribution | -0.10 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.23 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.38 |
| SVM RNA-class probability | 0.713029 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 20594761 93 - 27905053 AAA---AGAUAAAAUUCCAGCAUAUGUGUGUGGAUUUUUAGGA-ACAGAAUCCAUUGG----AGGUUCCAAAUAU--AUAUGUAAAGUGUGCGGAUGUCGUUA ...---.((((....(((.((((((((((.((((((((.....-..))))))))((((----.....))))))))--))))))...(....)))))))).... ( -22.60) >DroSec_CAF1 19564 98 - 1 AAA---AGAUAAAAGUCCAGCAUAUUAGUGUGGAUUUUUAGUAAACGGGAUCCAUUGGAUGGAGGUUCCGAAUAU--AUAUGUAAGGUGUGCUGAUGUCGUUA ...---...((((((((((.(((....)))))))))))))...(((((.(((..(((((.......)))))...(--((((.....)))))..))).))))). ( -22.80) >DroSim_CAF1 19678 102 - 1 AAAGAGAGAUAAAAGUCCAGCAUAUGUGUGUGGAUUUUUAGUA-ACAGGAUCCAUUGGAGGGAGGUUCCAAAUAUAUAUAUGUAAAAUGUACUGAUGUUGUUA .......(((((.(((.(((((((((((((((((((((.....-..))))))).((((((.....))))))))))))))))))....)).)))....))))). ( -24.20) >consensus AAA___AGAUAAAAGUCCAGCAUAUGUGUGUGGAUUUUUAGUA_ACAGGAUCCAUUGGA_GGAGGUUCCAAAUAU__AUAUGUAAAGUGUGCUGAUGUCGUUA .......((((..(((.((((((((..(((((((((((........))))))).(((((.......)))))))))..))))))....)).)))..)))).... (-16.23 = -16.13 + -0.10)



| Location | 20,594,798 – 20,594,892 |
|---|---|
| Length | 94 |
| Sequences | 4 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 76.12 |
| Mean single sequence MFE | -17.38 |
| Consensus MFE | -10.10 |
| Energy contribution | -10.10 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.80 |
| SVM RNA-class probability | 0.855134 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 20594798 94 + 27905053 CU-----CCAAUGGAUUCUGU-UCCU--------------AAAAAUCCACACACAUAUGCUGGAAUUUUAUCU---UUUUUUCGGUGUAGAGAAAUUUAUCAUUUCACUGCAGCACA ..-----.....(((......-)))(--------------((((.((((((......)).)))).)))))...---.......(.(((((.(((((.....))))).))))).)... ( -18.20) >DroSec_CAF1 19601 99 + 1 CUCCA-UCCAAUGGAUCCCGUUUACU--------------AAAAAUCCACACUAAUAUGCUGGACUUUUAUCU---UUUUUUCGGUGUAGAGAAAUUUAUCAUUUCACUGCAGCACA .((((-(...)))))..........(--------------((((.((((((......)).)))).)))))...---.......(.(((((.(((((.....))))).))))).)... ( -18.50) >DroSim_CAF1 19717 101 + 1 CUCCC-UCCAAUGGAUCCUGU-UACU--------------AAAAAUCCACACACAUAUGCUGGACUUUUAUCUCUCUUUUUUCGGUGUAGAGAAAUUUAUCAUUUCACUGCAGCACA ...((-......)).......-...(--------------((((.((((((......)).)))).))))).............(.(((((.(((((.....))))).))))).)... ( -16.90) >DroEre_CAF1 19523 104 + 1 CUCCAAA---------UCUGU-UACUUUUUAGUAAUAUUUAGUAACCCACCAACAUAUUUUGAAUUCUUCUCU---UUUUCUCGGUGUAGAGAAAUUUAUCAUUUCACUGCAGCACA .......---------...((-((((...((....))...))))))...........................---.......(.(((((.(((((.....))))).))))).)... ( -15.90) >consensus CUCCA_UCCAAUGGAUCCUGU_UACU______________AAAAAUCCACACACAUAUGCUGGACUUUUAUCU___UUUUUUCGGUGUAGAGAAAUUUAUCAUUUCACUGCAGCACA ...................................................................................(.(((((.(((((.....))))).))))).)... (-10.10 = -10.10 + -0.00)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:51:49 2006