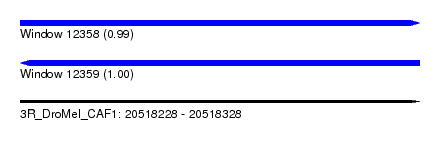
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 20,518,228 – 20,518,328 |
| Length | 100 |
| Max. P | 0.996789 |

| Location | 20,518,228 – 20,518,328 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 72.79 |
| Mean single sequence MFE | -33.67 |
| Consensus MFE | -17.98 |
| Energy contribution | -18.88 |
| Covariance contribution | 0.91 |
| Combinations/Pair | 1.21 |
| Mean z-score | -5.56 |
| Structure conservation index | 0.53 |
| SVM decision value | 2.23 |
| SVM RNA-class probability | 0.990798 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 20518228 100 + 27905053 -------------UCGGU--CUACAAACUGAAGCUUAAUUGUGAUUUAUGGAUGUGUAUUUUUUAGCAGUUAAAAAUCACAAUUUAGCCACU-UUGUAG-GUCGAUGAAACAGCUCA -------------(((..--(((((((.((..(((.(((((((((((.(((.(((..........))).))).))))))))))).))))).)-))))))-..)))............ ( -31.40) >DroSec_CAF1 720 100 + 1 -------------UCGGU--CUACAAACUGAAGCUUAAUUGUGAUUUAAGGAUGUGUAUUUUUUUGCACUAAAAAAUCACAAUUUAGCCAAU-UUGUAG-GUCGAUGAAAAACCACA -------------(((..--(((((((.((..(((.(((((((((((......(((((......)))))....))))))))))).))))).)-))))))-..)))............ ( -34.40) >DroSim_CAF1 740 99 + 1 -------------UCGGU--CUACAAACUGAAGCUUAAUUGUGAUUUAAGGAUGUGUAUUUUUUAGCACUAAAAAAUCACAAUUUAGCCAAU-UUGUAG-GUCGAU-AAAAACCACA -------------(((..--(((((((.((..(((.(((((((((((......((((........))))....))))))))))).))))).)-))))))-..))).-.......... ( -32.60) >DroEre_CAF1 742 99 + 1 -------------UCGGC--CUACAAACAUAAGCAUAAUUGUGAUUUAAGGAUGUAUAUUU-CAAGUAGUAAUACAUCACAAUUUAGCCAAU-UUGUAG-GCCGAUGAAAAACCUCA -------------(((((--(((((((.....((..(((((((((....(((.......))-)..(((....))))))))))))..))...)-))))))-)))))............ ( -32.00) >DroYak_CAF1 789 100 + 1 -------------UCGGC--AUACAAACUUGAGCUUAAUUGUGAUUUAAGGAUGUGAAUUU-CAAGCAACGAAAAAUCACAAUGAAGCCAAUUUUGUAG-GCCGAUGGAACACCUCA -------------(((((--.((((((.(((.((((.((((((((((..(..(((......-...))).)...)))))))))).))))))).)))))).-)))))............ ( -32.40) >DroPer_CAF1 831 108 + 1 UACACACGUUUCUCUGGUUUAUACAAAAUGAAGUUUCAUUGUGAUUGGAGCCUA--UAUGUUCUGGGAUAAUCCAAUCACAAUCAAGCUUC--CUGUACAACCCAUGAGAG-----A ........(((((((((.((.((((....(((((((.((((((((((((.((((--.......))))....)))))))))))).)))))))--.)))).)).))).)))))-----) ( -39.20) >consensus _____________UCGGU__CUACAAACUGAAGCUUAAUUGUGAUUUAAGGAUGUGUAUUUUCUAGCACUAAAAAAUCACAAUUUAGCCAAU_UUGUAG_GCCGAUGAAAAACCUCA .............((((((.((((((......(((.(((((((((((.....(((..........))).....))))))))))).))).....))))))))))))............ (-17.98 = -18.88 + 0.91)



| Location | 20,518,228 – 20,518,328 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 72.79 |
| Mean single sequence MFE | -30.47 |
| Consensus MFE | -17.95 |
| Energy contribution | -18.20 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.21 |
| Mean z-score | -4.50 |
| Structure conservation index | 0.59 |
| SVM decision value | 2.75 |
| SVM RNA-class probability | 0.996789 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 20518228 100 - 27905053 UGAGCUGUUUCAUCGAC-CUACAA-AGUGGCUAAAUUGUGAUUUUUAACUGCUAAAAAAUACACAUCCAUAAAUCACAAUUAAGCUUCAGUUUGUAG--ACCGA------------- ............(((..-((((((-(.(((((.(((((((((((.....((............)).....))))))))))).)))..)).)))))))--..)))------------- ( -24.60) >DroSec_CAF1 720 100 - 1 UGUGGUUUUUCAUCGAC-CUACAA-AUUGGCUAAAUUGUGAUUUUUUAGUGCAAAAAAAUACACAUCCUUAAAUCACAAUUAAGCUUCAGUUUGUAG--ACCGA------------- ............(((..-((((((-(((((((.(((((((((((....(((..........)))......))))))))))).)))..))))))))))--..)))------------- ( -28.30) >DroSim_CAF1 740 99 - 1 UGUGGUUUUU-AUCGAC-CUACAA-AUUGGCUAAAUUGUGAUUUUUUAGUGCUAAAAAAUACACAUCCUUAAAUCACAAUUAAGCUUCAGUUUGUAG--ACCGA------------- ..........-.(((..-((((((-(((((((.(((((((((((....(((..........)))......))))))))))).)))..))))))))))--..)))------------- ( -28.30) >DroEre_CAF1 742 99 - 1 UGAGGUUUUUCAUCGGC-CUACAA-AUUGGCUAAAUUGUGAUGUAUUACUACUUG-AAAUAUACAUCCUUAAAUCACAAUUAUGCUUAUGUUUGUAG--GCCGA------------- ............(((((-((((((-((.(((..((((((((((((((.(.....)-.)))))..........)))))))))..)))...))))))))--)))))------------- ( -33.00) >DroYak_CAF1 789 100 - 1 UGAGGUGUUCCAUCGGC-CUACAAAAUUGGCUUCAUUGUGAUUUUUCGUUGCUUG-AAAUUCACAUCCUUAAAUCACAAUUAAGCUCAAGUUUGUAU--GCCGA------------- ...((....)).(((((-.((((((.(((((((.((((((((((.....((..((-.....)))).....)))))))))).)))).))).)))))).--)))))------------- ( -32.80) >DroPer_CAF1 831 108 - 1 U-----CUCUCAUGGGUUGUACAG--GAAGCUUGAUUGUGAUUGGAUUAUCCCAGAACAUA--UAGGCUCCAAUCACAAUGAAACUUCAUUUUGUAUAAACCAGAGAAACGUGUGUA .-----.((((.(((.(((((((.--((((.((.((((((((((((....((.........--..)).)))))))))))).)).))))....))))))).))))))).......... ( -35.80) >consensus UGAGGUUUUUCAUCGAC_CUACAA_AUUGGCUAAAUUGUGAUUUUUUACUGCUAAAAAAUACACAUCCUUAAAUCACAAUUAAGCUUCAGUUUGUAG__ACCGA_____________ ............(((...((((((....((((.(((((((((((..........................))))))))))).)))).....))))))....)))............. (-17.95 = -18.20 + 0.25)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:51:21 2006