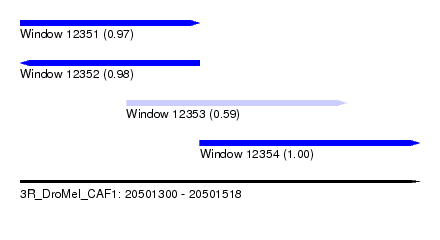
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 20,501,300 – 20,501,518 |
| Length | 218 |
| Max. P | 0.999087 |

| Location | 20,501,300 – 20,501,398 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 75.03 |
| Mean single sequence MFE | -26.22 |
| Consensus MFE | -18.50 |
| Energy contribution | -18.50 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.71 |
| SVM decision value | 1.61 |
| SVM RNA-class probability | 0.967206 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
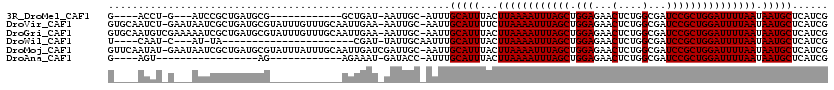
>3R_DroMel_CAF1 20501300 98 + 27905053 G----ACCU-G---AUCCGCUGAUGCG------------GCUGAU-AAUUGC-AUUUGCAUUUACUUAAAAUUUAGCUGGAGAACUCUGGCGAUCCGCUGGAUUUUAAUAAUGCUCAUCG .----....-(---((((((....)))------------).....-......-....(((((...((((((((((((.(((...(....)...))))))))))))))).)))))..))). ( -25.00) >DroVir_CAF1 57186 117 + 1 GUGCAAUCU-GAAUAAUCGCUGAUGCGUAUUUGUUUGCAAUUGAA-AAUUGC-AAUUGCAUUUUCUUAAAAUUUAGCUGGAGAACUCUGGCGAUCCGCUGGAUUUUAAUAAUGCUCAUCG (.(((....-((((((.(((....)))...))))))(((((((..-.....)-))))))......((((((((((((.(((...(....)...)))))))))))))))...))).).... ( -30.20) >DroGri_CAF1 67325 118 + 1 GUGCAAUGUCGAAAAAUCGCUGAUGCGUAUUUGUUUGCAAUUGAA-AAUUGC-AAUUGCAUUUACUUAAAAUUUAGCUGGAGAACUCUGGCGAUCCGCUGGAUUUUAAUAAUGCUCAUCG ..((.((........)).)).((((.(((((.((.((((((((..-.....)-)))))))...))((((((((((((.(((...(....)...))))))))))))))).))))).)))). ( -30.50) >DroWil_CAF1 56983 88 + 1 U----CAAU-C---AU-UA----------------------CGAU-UAUUGCAAUUUGCAUUUACUUAAAAUUUAGCUGGAGAACUCUGGCGAUCCGCUGGAUUUUAAUAAUGCUCAUCG .----.(((-(---..-..----------------------.)))-)..........(((((...((((((((((((.(((...(....)...))))))))))))))).)))))...... ( -19.30) >DroMoj_CAF1 63312 118 + 1 GUUCAAUAU-GAAUAAUCGCUGAUGCGUAUUUAUUUGCAAUUGAUCGAUUGC-AAUUGCAUUUACUUAAAAUUUAGCUGGAGAACUCUGGCGAUCCGCUGGAUUUUAAUAAUGCUCAUCG ((((.....-)))).......((((.(((((((..((((((((........)-)))))))..)).((((((((((((.(((...(....)...))))))))))))))).))))).)))). ( -31.70) >DroAna_CAF1 50473 85 + 1 G----AGU-----------------AG------------AGAAAU-GAUACC-AUUUGCAUUUACUUAAAAUUUAGCUGGAGAACUCUGGCGAUCCGCUGGAUUUUAAUAAUGCUCAUCG (----(((-----------------(.------------..((((-(....)-))))........((((((((((((.(((...(....)...)))))))))))))))...))))).... ( -20.60) >consensus G____AUAU_G___AAUCGCUGAUGCG____________AUUGAU_AAUUGC_AAUUGCAUUUACUUAAAAUUUAGCUGGAGAACUCUGGCGAUCCGCUGGAUUUUAAUAAUGCUCAUCG .........................................................(((((...((((((((((((.(((...(....)...))))))))))))))).)))))...... (-18.50 = -18.50 + 0.00)

| Location | 20,501,300 – 20,501,398 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 75.03 |
| Mean single sequence MFE | -23.82 |
| Consensus MFE | -15.30 |
| Energy contribution | -15.30 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.15 |
| Structure conservation index | 0.64 |
| SVM decision value | 1.90 |
| SVM RNA-class probability | 0.981914 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 20501300 98 - 27905053 CGAUGAGCAUUAUUAAAAUCCAGCGGAUCGCCAGAGUUCUCCAGCUAAAUUUUAAGUAAAUGCAAAU-GCAAUU-AUCAGC------------CGCAUCAGCGGAU---C-AGGU----C .(((..(((((.(((((((..((((((..((....))..))).)))..)))))))...)))))....-......-.....(------------(((....))))))---)-....----. ( -23.10) >DroVir_CAF1 57186 117 - 1 CGAUGAGCAUUAUUAAAAUCCAGCGGAUCGCCAGAGUUCUCCAGCUAAAUUUUAAGAAAAUGCAAUU-GCAAUU-UUCAAUUGCAAACAAAUACGCAUCAGCGAUUAUUC-AGAUUGCAC .((((.(((((.(((((((..((((((..((....))..))).)))..)))))))...)))))..((-(((((.-....))))))).........)))).((((((....-.)))))).. ( -30.70) >DroGri_CAF1 67325 118 - 1 CGAUGAGCAUUAUUAAAAUCCAGCGGAUCGCCAGAGUUCUCCAGCUAAAUUUUAAGUAAAUGCAAUU-GCAAUU-UUCAAUUGCAAACAAAUACGCAUCAGCGAUUUUUCGACAUUGCAC .((((.(((((.(((((((..((((((..((....))..))).)))..)))))))...)))))..((-(((((.-....))))))).........)))).(((((........))))).. ( -29.00) >DroWil_CAF1 56983 88 - 1 CGAUGAGCAUUAUUAAAAUCCAGCGGAUCGCCAGAGUUCUCCAGCUAAAUUUUAAGUAAAUGCAAAUUGCAAUA-AUCG----------------------UA-AU---G-AUUG----A ((((..(((((.(((((((..((((((..((....))..))).)))..)))))))...)))))..))))...((-(((.----------------------..-..---)-))))----. ( -18.10) >DroMoj_CAF1 63312 118 - 1 CGAUGAGCAUUAUUAAAAUCCAGCGGAUCGCCAGAGUUCUCCAGCUAAAUUUUAAGUAAAUGCAAUU-GCAAUCGAUCAAUUGCAAAUAAAUACGCAUCAGCGAUUAUUC-AUAUUGAAC ..(((((((((.(((((((..((((((..((....))..))).)))..)))))))...)))))..((-(((((......))))))).......(((....))).....))-))....... ( -26.20) >DroAna_CAF1 50473 85 - 1 CGAUGAGCAUUAUUAAAAUCCAGCGGAUCGCCAGAGUUCUCCAGCUAAAUUUUAAGUAAAUGCAAAU-GGUAUC-AUUUCU------------CU-----------------ACU----C ......(((((.(((((((..((((((..((....))..))).)))..)))))))...)))))((((-(....)-))))..------------..-----------------...----. ( -15.80) >consensus CGAUGAGCAUUAUUAAAAUCCAGCGGAUCGCCAGAGUUCUCCAGCUAAAUUUUAAGUAAAUGCAAAU_GCAAUU_AUCAAU____________CGCAUCAGCGAUU___C_AGAU____C ......(((((.(((((((..((((((..((....))..))).)))..)))))))...)))))......................................................... (-15.30 = -15.30 + -0.00)


| Location | 20,501,358 – 20,501,478 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 99.11 |
| Mean single sequence MFE | -31.38 |
| Consensus MFE | -30.53 |
| Energy contribution | -30.53 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -0.74 |
| Structure conservation index | 0.97 |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.590284 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 20501358 120 + 27905053 AGAACUCUGGCGAUCCGCUGGAUUUUAAUAAUGCUCAUCGUUUAUCGUAUUGGACCUGUGUCUAUUUCCUAAUUGUGACCAUGUCAACGGUAGGAUAUGGUGACGUUUACUGUGAGACUG .(((.((..(((...)))..)).))).......(((((.((..((.....(((((....)))))..........((.((((((((........)))))))).))))..)).))))).... ( -30.50) >DroVir_CAF1 57263 120 + 1 AGAACUCUGGCGAUCCGCUGGAUUUUAAUAAUGCUCAUCGUUUAUCCUAUUGGACCUGUGUCUAUUUCCUAAUUGUGACCAUGUCAACGGUAGGAUAUGGUGACGUUUACUGUGAGACUG .....((..(((...)))..))(((((..((((.(((((((..((((((((((((.((.(((.(((....)))...))))).)))..)))))))))))))))))))).....)))))... ( -32.20) >DroGri_CAF1 67403 120 + 1 AGAACUCUGGCGAUCCGCUGGAUUUUAAUAAUGCUCAUCGUUUAUCCUAUUGGACCUGUGUCUAUUUCCUAAUUGUGACCAUGUCAACGGUAGGAUAUGGUGACGUUUACUGUGAGACUG .....((..(((...)))..))(((((..((((.(((((((..((((((((((((.((.(((.(((....)))...))))).)))..)))))))))))))))))))).....)))))... ( -32.20) >DroWil_CAF1 57031 120 + 1 AGAACUCUGGCGAUCCGCUGGAUUUUAAUAAUGCUCAUCGUUUAUCCUAUUGGACCUGUGUCUAUUUCCUAAUUGUGACCAUGUCAACGGUAGGAUAUGGUGACGUUUACUGUGAGACUG .....((..(((...)))..))(((((..((((.(((((((..((((((((((((.((.(((.(((....)))...))))).)))..)))))))))))))))))))).....)))))... ( -32.20) >DroMoj_CAF1 63390 120 + 1 AGAACUCUGGCGAUCCGCUGGAUUUUAAUAAUGCUCAUCGUUUAUCUUAUUGGACCUGUGUCUAUUUCCUAAUUGUGACCAUGUCGACGGUAGGAUAUGGUGACGUUUACUGUGAGACUG .(((.((..(((...)))..)).))).......(((((.((..((.....(((((....)))))..........((.((((((((........)))))))).))))..)).))))).... ( -30.70) >DroAna_CAF1 50518 120 + 1 AGAACUCUGGCGAUCCGCUGGAUUUUAAUAAUGCUCAUCGUUUAUCGUAUUGGACCUGUGUCUAUUUCCUAAUUGUGACCAUGUCAACGGUAGGAUAUGGUGACGUUUACUGUGAGACUG .(((.((..(((...)))..)).))).......(((((.((..((.....(((((....)))))..........((.((((((((........)))))))).))))..)).))))).... ( -30.50) >consensus AGAACUCUGGCGAUCCGCUGGAUUUUAAUAAUGCUCAUCGUUUAUCCUAUUGGACCUGUGUCUAUUUCCUAAUUGUGACCAUGUCAACGGUAGGAUAUGGUGACGUUUACUGUGAGACUG .(((.((..(((...)))..)).))).......(((((.((..((.....(((((....)))))..........((.((((((((........)))))))).))))..)).))))).... (-30.53 = -30.53 + 0.00)



| Location | 20,501,398 – 20,501,518 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.28 |
| Mean single sequence MFE | -34.65 |
| Consensus MFE | -34.36 |
| Energy contribution | -34.08 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.99 |
| SVM decision value | 3.36 |
| SVM RNA-class probability | 0.999087 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 20501398 120 + 27905053 UUUAUCGUAUUGGACCUGUGUCUAUUUCCUAAUUGUGACCAUGUCAACGGUAGGAUAUGGUGACGUUUACUGUGAGACUGUCCUGGGAAGAACAUUUCUCGUGUUCUUUCUGCUCGUCGG ...........((((....((((......(((.(((.((((((((........)))))))).))).))).....)))).)))).(((((((((((.....)))))))))))......... ( -35.40) >DroVir_CAF1 57303 120 + 1 UUUAUCCUAUUGGACCUGUGUCUAUUUCCUAAUUGUGACCAUGUCAACGGUAGGAUAUGGUGACGUUUACUGUGAGACUGUUUUGGGAAGAACAUUUCUCGUGUUCUUUCUGCUCGUCGG ............(((..((((((......(((.(((.((((((((........)))))))).))).))).....))))......(((((((((((.....)))))))))))))..))).. ( -34.10) >DroGri_CAF1 67443 120 + 1 UUUAUCCUAUUGGACCUGUGUCUAUUUCCUAAUUGUGACCAUGUCAACGGUAGGAUAUGGUGACGUUUACUGUGAGACUGUCUUAGGGAGAACAUUUCUCGUGUUCUUUCUGCUCGUCGG ..(((((((((((((.((.(((.(((....)))...))))).)))..))))))))))((....))....(((((((.......(((..(((((((.....)))))))..))))))).))) ( -34.51) >DroWil_CAF1 57071 120 + 1 UUUAUCCUAUUGGACCUGUGUCUAUUUCCUAAUUGUGACCAUGUCAACGGUAGGAUAUGGUGACGUUUACUGUGAGACUGUACUGGGAAGAACAUUUCUCGUGUUCUUUCUGCUUGUCGG ............(((..((((((......(((.(((.((((((((........)))))))).))).))).....))))......(((((((((((.....)))))))))))))..))).. ( -34.20) >DroMoj_CAF1 63430 120 + 1 UUUAUCUUAUUGGACCUGUGUCUAUUUCCUAAUUGUGACCAUGUCGACGGUAGGAUAUGGUGACGUUUACUGUGAGACUGUCUUGGGAAGAACAUUUCUCGUGUUCUUUCUGCUCGUCGG ............(((..((((((......(((.(((.((((((((........)))))))).))).))).....))))......(((((((((((.....)))))))))))))..))).. ( -34.30) >DroAna_CAF1 50558 120 + 1 UUUAUCGUAUUGGACCUGUGUCUAUUUCCUAAUUGUGACCAUGUCAACGGUAGGAUAUGGUGACGUUUACUGUGAGACUGUCCUGGGAAGAACAUUUCUCGUGUUCUUUCUGCUCGUCGG ...........((((....((((......(((.(((.((((((((........)))))))).))).))).....)))).)))).(((((((((((.....)))))))))))......... ( -35.40) >consensus UUUAUCCUAUUGGACCUGUGUCUAUUUCCUAAUUGUGACCAUGUCAACGGUAGGAUAUGGUGACGUUUACUGUGAGACUGUCCUGGGAAGAACAUUUCUCGUGUUCUUUCUGCUCGUCGG ............(((..((((((......(((.(((.((((((((........)))))))).))).))).....))))......(((((((((((.....)))))))))))))..))).. (-34.36 = -34.08 + -0.28)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:51:16 2006